4.3 Rethinking the U-Net architecture for multimodal biomedical image segmentation
Grudzień Adrianna, Łukaszyk Marcin, Piasecki Michał
4.3.1 Abstrac
In our work we wanted to explore what reproducibility is, and how it applies to deep learning. The articles we had choesen are An Improvement of Data Classification Using Random Multimodel Deep Learning (RMDL) and MultiResUNet : Rethinking the U-Net architecture for multimodal biomedical image segmentation. In both papers we faced simmilar issues and limitation ,yet we have concluded our work with diffrent results for each article. First paper is unreproducible and second one is mostly reproducible.
4.3.2 What Reproducibility Is?
Reproducibility of an experiment plays crucial role in scientific community and serves as a proof for validity of both experiment and its consequences. It constitutes a basis of scientific endeavours and enable us to use science as a method to comprehend reality around us.
This essential term means ability to repeat particular experiment and obtain similar results, while having the same conditions, data, tools and means of measurement. Unfortunately, it is not always possible to repeat experiments and conditions required, them might prevent larger community to conduct them. To show these difficulties, we would like to provide two examples, which clearly illustrate, that some experiments might be almost impossible to repeat.
Firstly, let’s consider repeating Galileo experiments of dropping objects from fixed height to establish, whether objects fall with the same velocity. Such simple test is available for everyone, who is eager to explore experimentally basic Newtonian principles. On the other hand, if we would like to switch to quantum physics and prove experimentally quantum nonlocality, we are more than likely to be unable to conduct experiments proving this theory. The infrastructure required to conduct such experiments makes reproducibility an exclusive idea limited to only handful of people.
It is hard to underestimate the significance of reproducibility in science. We listed below main reasons for its importance:
1 Proof for correctness of experiment
Having obtained same results of an experiment conducted many times by the independent researchers, we are ensured that results are not coincidence. As a result, we can start treating new results as a proper description of phenomena around us.
2 Different views on experiment
After repeating an experiment and acquiring same results, other researchers may draw different, interesting conclusions. It enables to use conducted experiment to full extent, squeezing out as many conclusions about reality as it is possible.
3 Possibility of improving experiment
Other researchers may figure out how to improve experiment to obtain results in more efficient or precise way.
4.3.3 First article (An Improvement of Data Classification Using Random Multimodel Deep Learning (RMDL) )
4.3.3.1 Summary of Article
The main purpose of the article is creating combinations of different neural networks and finding the best model. In order to achieve these goal, authors use various optimalization methods , dropout layers and different models for prediction.
Authors describe methods used in article: TF-IDF, Word2Vec for Feature Extraction, NaïveBayesClassifier, SVM and S2GD for classification and types of neural networks : Deep Neural Network, Convolutional Neural Network and Recurrent Neural Network. They explain all algorithms and ideas used in their work. Consequently, they combine different architectures to obtain one model – RMDL. Having constructed the model, authors decide to evaluate their masterpiece on different datasets. They use it both on text data (WOS, Reuters IMDB, 20newsgroup) and image data (MNIST, Cifar-10). Authors evaluate model with metrics like : Precision, Recall F1 and Score. The all model is build with usage of Cuda, tensorflow and keras libraries.
At the end, authors discuss how its model might come useful in classification task.
4.3.3.2 Our Work with Reproducibility of First Article
We had run all our experiments in google Collaboratory.
One of the datasets was not available due to unavailability of server that article was referring to. Rest of datasets were freely accessible. Most of our issues were due to hardware limitations. The predominant ones were RAM lack of free memory errors and memory leaks. Collaboratory doesn’t allow to run program more than 12 hours straight so we couldn’t train the biggest models.
Our main goal was to explore reproducibility of given paper, and main issue that we combined were lack of definitions of terms that authors used. One of them was error rate that wasn’t defined. They didn’t specify what depth of RMDL is and a lot of our work was to guess based of their results what is the proper definition.
4.3.3.3 Reproducibility of this Article
We cannot acknowledge it as reproducible. The results that were achieved by authors are not properly documented and defined in article making it hard to check if we had similar results. Some of our struggle results from the hardware limitations where we cannot train similar sized models or in extreme situations cannot train at all.
4.3.4 Second article (MultiResUNet : Rethinking the U-Net architecture for multimodal biomedical image segmentation)
4.3.4.1 Introduction
Throughout XX century we have developed multiple methods for imaging human bodies. Radiography, functional magnetic resonance and fluoroscopia are just mere examples of methods we invented. It is hard to underestimate its benefits, they enable us to understand processes inside our bodies to much greater extent. Thanks to them, it is possible to detect pathologies and monitor our health at scale not imaginable before. Having been exposed to such vast amount of medical images, computer scientists started thinking about creating software, which could analyse them automatically. Since late 60’s researchers have been attempting to build models segmenting medical images and conducting diagnosis. Were we to create such software, we would relieve doctors from segmenting images manually and support them with independent diagnosis from computer. As a result, it would make our healthcare system less overwhelmed and more people could seek help in predicament.
4.3.4.2 Summary
There were multiple ways of creating such models. Firstly, ‘rule-based’ approach was commonplace, with scientists explicitly setting rules for model evaluation. Poor generalization resulted in different approaches based on geometrical analysis or fuzzy logic. Nonetheless, recently we can notice general shift towards Deep Learning as a way of segmenting and diagnosing images. Convolutional neural networks have been obtaining outstanding results, with U-Net architecture standing out of the crowd.
We would like to review article “MultiResUNet Rethinking the U-Net Architecture for biomedical image segmentation.” In this article, modification of U-Net – MultiResUNet is proposed. Authors are convinced that small changes in well know U-Net architecture lead to better model performance. Authors list situations, in which U-Net architecture do not get sufficient results.
4.3.4.3 What is U-Net?
U-net is a deep learning architecture that was proposed in 2015. It’s a modification of simple neural network with odd number layers of convolution layers and a max-pooling or up-convolutions layers, which is used to for segmentation of images. This modification is a series of connections between each simmilar distanced layer from middle one. This “bends” our network making it U-shaped, thus the name. Each level of network consists of series of convolution layers and then max-pool or up-convolution layer that depends on which side of and U shape are we in.
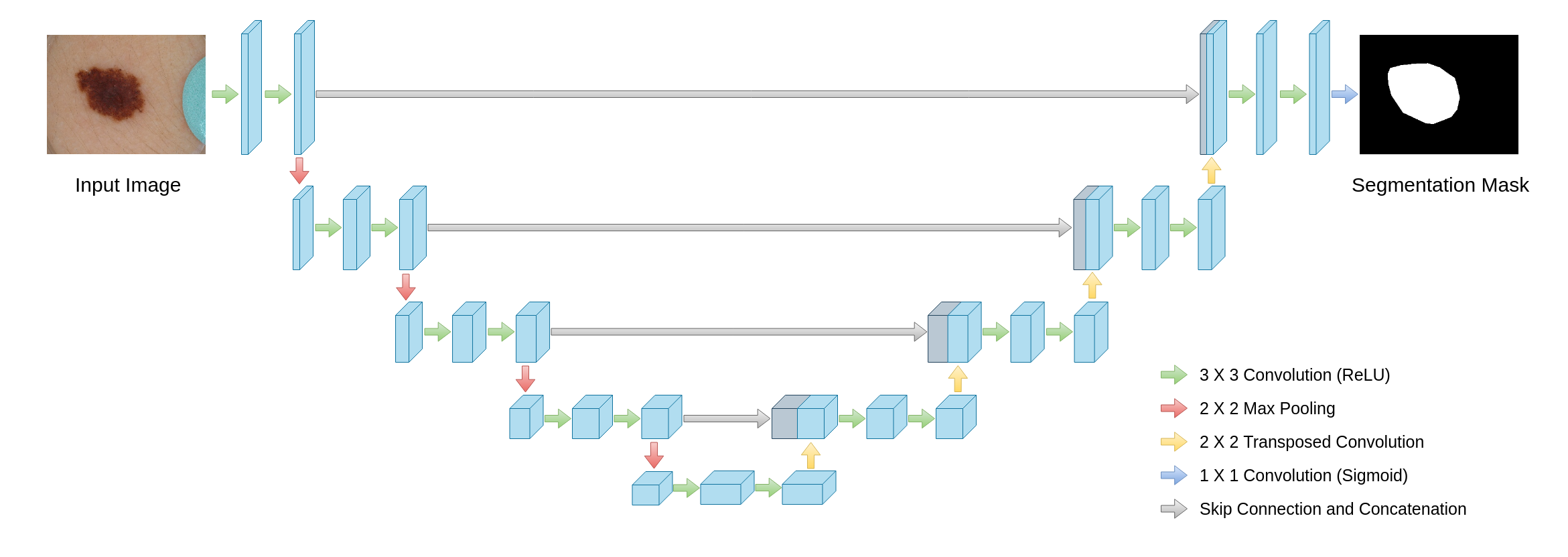
Figure 4.1: Example of U-net
In order to improve U-Net , authors decide to introduce following changes in model architecture. They factorize the bigger and more expensive 5 × 5 and 7 × 7 filters as a succession of 3 × 3 filters. This saves computations as we need calculate 3 3x3 convolutions layers to have similar results as calculating 3x3, 5x5 and 7x7 layers. They concatenate all of the estimated layers with one made from 1x1 convolution layers. This allows to better retrieve spatial features from different scales.
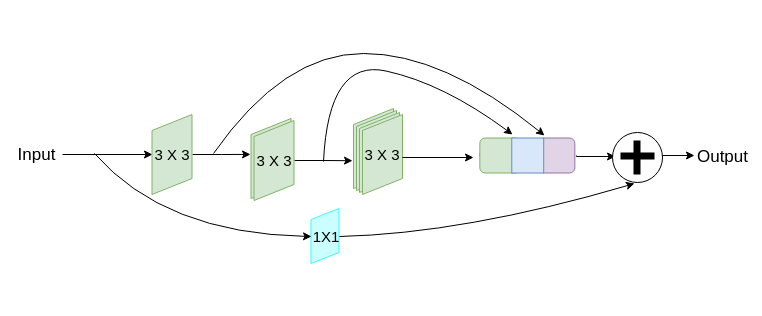
Figure 4.2: Example of ResBlock
The excellence of U-Net architecture stems mainly from the introduction of connection between layers, which enables to save spatial information while going deeper into U-Net architecture. However, authors speculate that combination of a simple copy of an output of previous convolutional layer and information received after all transformations conducted in model results in discrepancy in information carried by both of them. To minimize this discrepancy authors suggest “ResPath” - series of consecutive convolutional layers. They decrease number of convolutional layers while going deeper into the architecture, due to smaller difference in information in “depth” of the model.
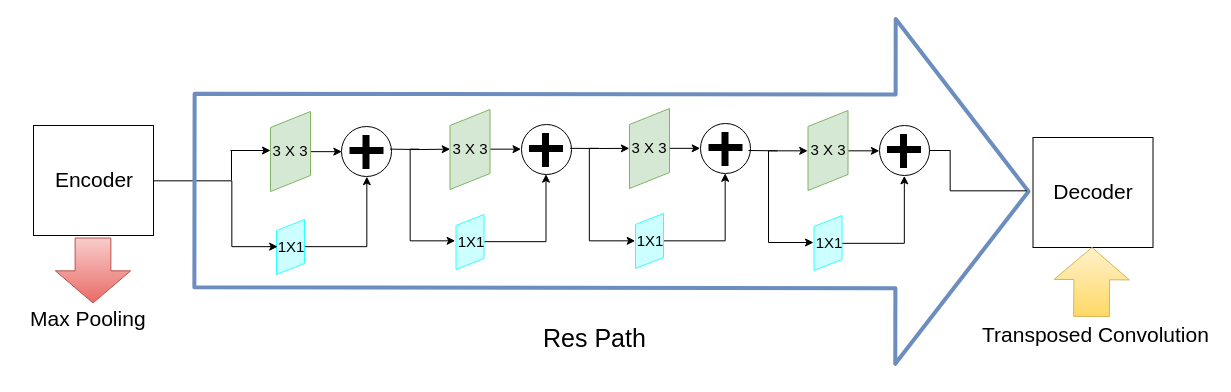
Figure 4.3: Example of ResPath
In the end we get simmilar looking neural network but with improvements in most places. Authors states that their structure has simmilar computational complexity but gives better results.
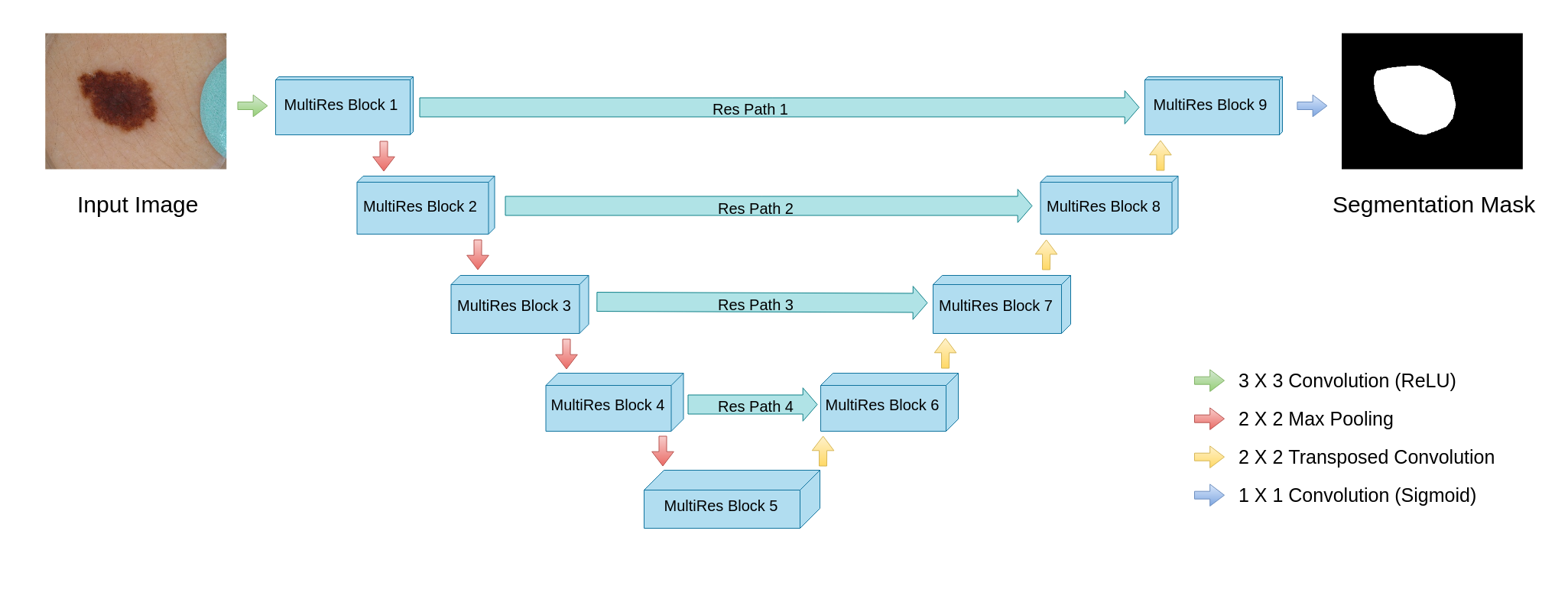
Figure 4.4: MultiResUNet
Authors test their improved architecture on 5 different datasets and compare results with “basic” U-Net. They introduce new metric: Jacard Index to measure performance of both models. For mask A and model’s marked area B it calculates the intersection of both sets divided by their union. Authors show that their model obtain better results, learns faster and is less vulnerable to perturbations.
4.3.4.4 Our Work with Reproducibility of Second Article
We decided to repeat experiments in Google Colab using Python
Unfortunately,we were not able to obtain all datasets used by authors. After long research, we managed to download datasets: Fluorescence microscopy images Murphy Lab, Electron microscopy images, Dermoscopy images IC-2017dataset
Having downloaded datasets, we encountered many problems. There were multiple problems with transforming datasets into desired format used by authors. Problems with conversion, partition of datasets and ambiguous files led to frustration. Moreover, Google Colab, environment in which we tried to reproduce results, had not enough RAM to smoothly generate results. At the end, we managed to obtain some results, which were similar to results from the article.
[Wykresy dermoskopi i endoskopi](#fig:Original Rsults)Orginal Plots
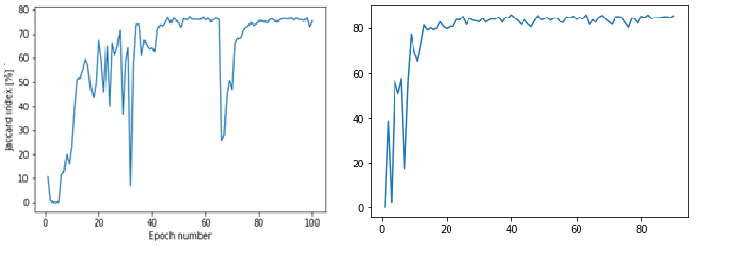
(#fig:Our Rsults)Our Plots
4.3.4.5 Reproducibility of this article
Article is reproducible. In article authors try to improve U-Net architecture. Providing simple explanations and empirical data, they explain motivation behind their improvements.
Unfortunately, few of datasets used by authors are no longer available, which may make it harder to conduct same experiments. However, rest of the datasets is easily accessible, so it is still possible to verify results obtained by authors.
When it comes to authors argumentation behind new architecture, we are not completely satisfied. As students, who have little expertise in using Machine Learning in medical areas, we are not satisfied with mathematical explanations provided by the authors. No mathematical proofs and small number of datasets make us feel unease, if authors’ architecture outperforms basic U-Net architecture globally.
4.3.5 Conclusion
In both articles we coudn’t simply state if article are reproducible. We had to have non-binary measure for each.
First article is mostly unreproducible due to lack of proper deffinitions thath dosn’t allow us to be sure thath our experiments are conducted in the same environment.
Second article is reproducible. Our tries of recreating simmilar results were mostly positive and most of our issues were with hardwere limitations. The biggest truble is some missing data stes thath are not longer available from refer to sources. Some of our worry is in how authors explained their reasoning as it’s mostly based on assumption and results.