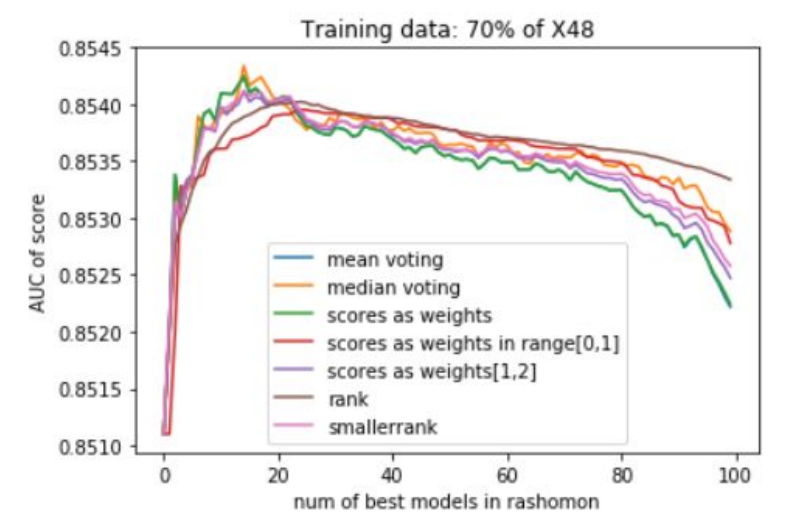
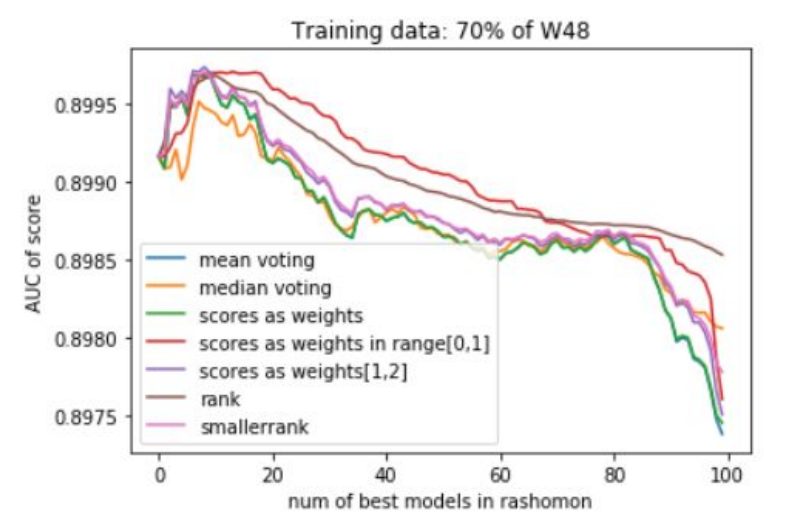
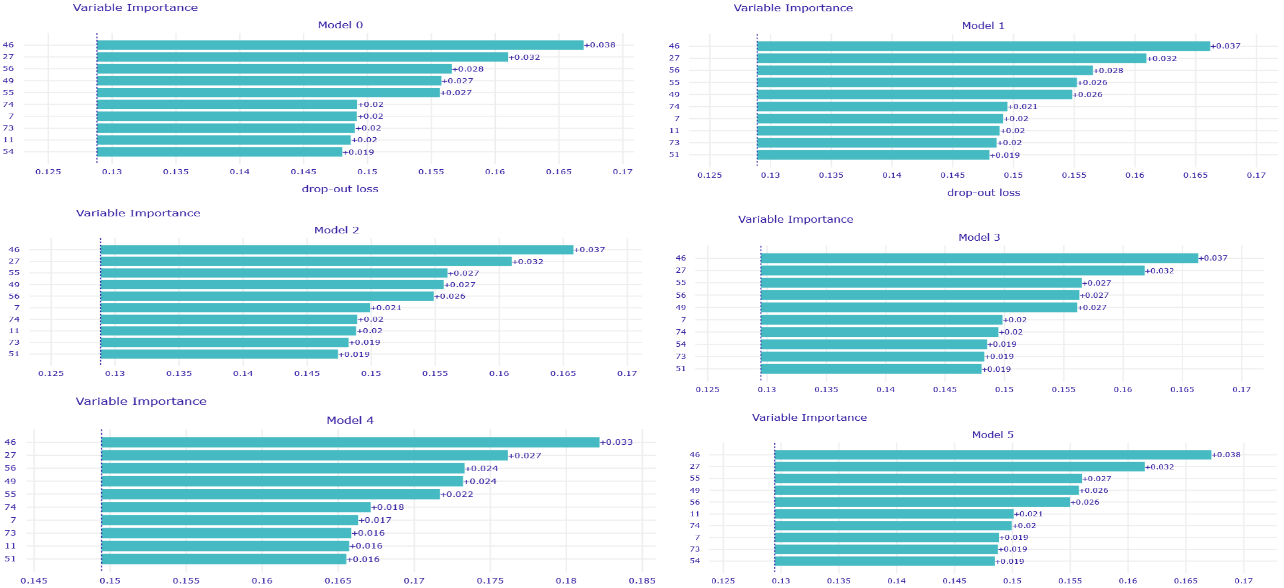
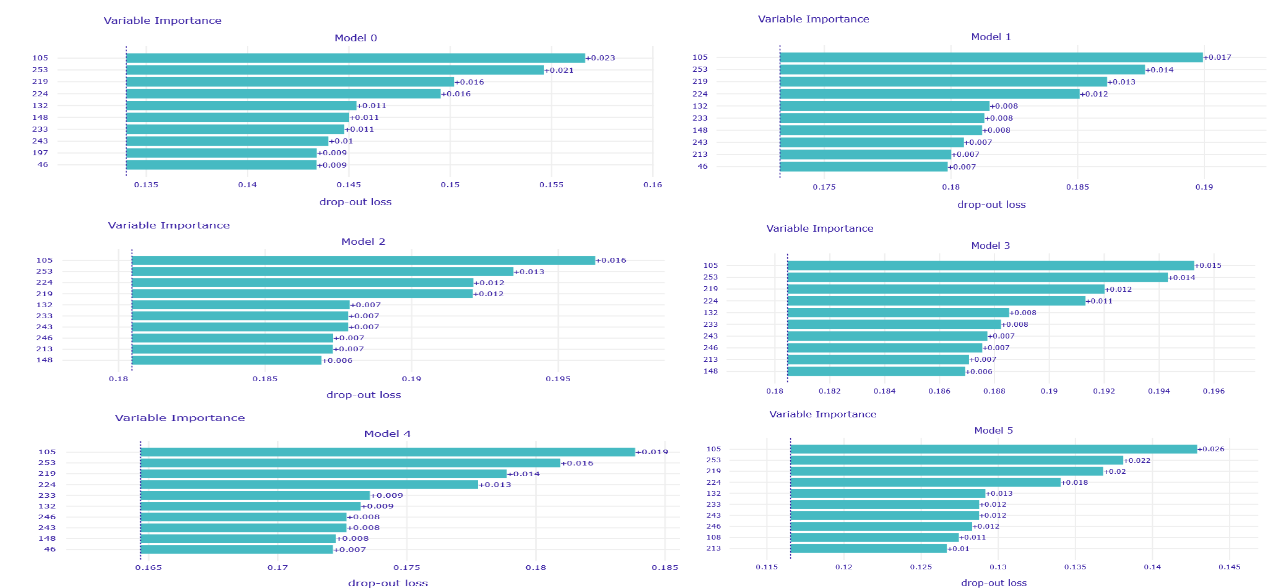
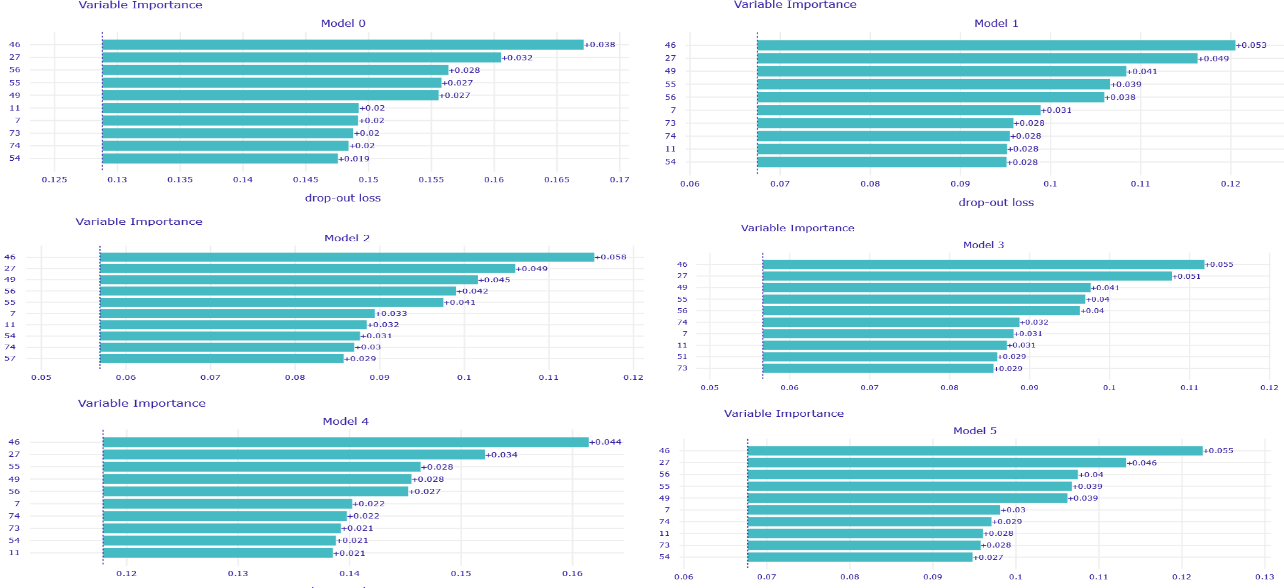
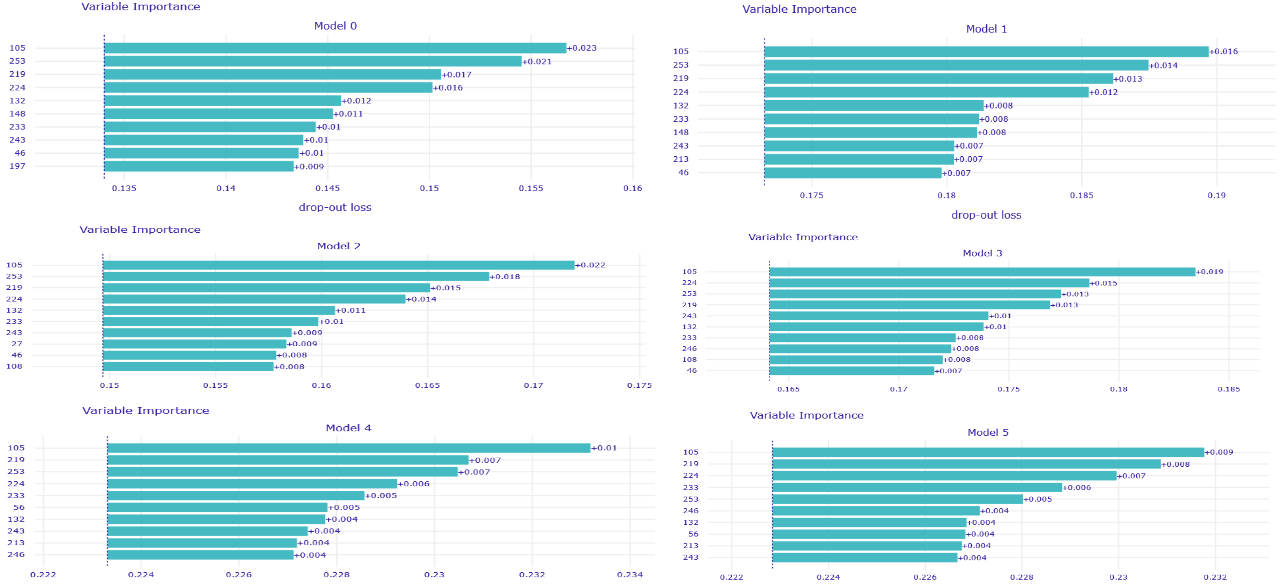
In addition to that, several voting strategies of creating Rashomon sets for further predictions were tried out. During this whole work, the set of most important features of MIMIC-III for the mortality prediction task was discovered, which also may be useful for further researches or could give rise to new medical conclusions.
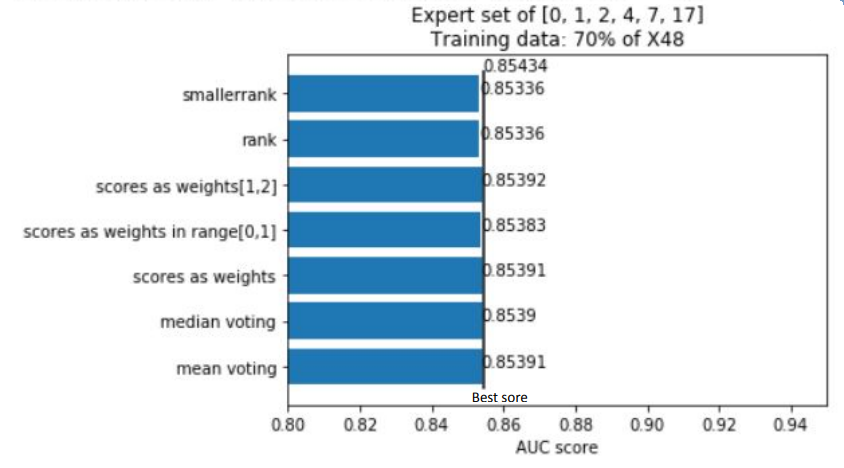
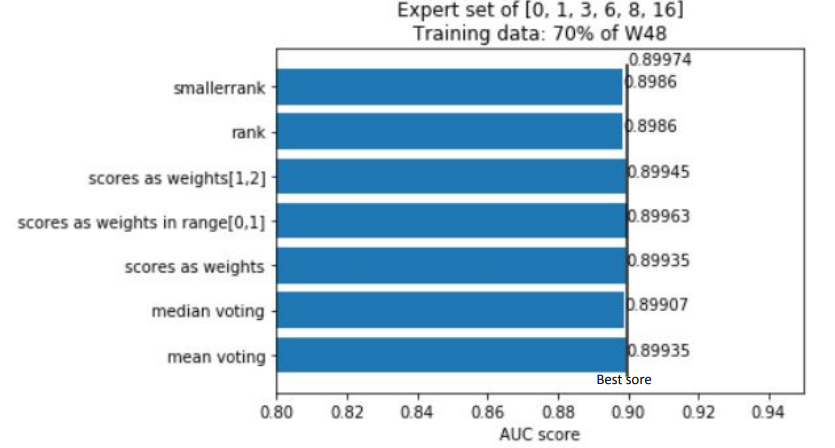
Summing the results of all experiments up, one can conclude that Rashomon expert sets are worth the attention of researchers even though in this study they have slightly underperformed top performance model sets. Because of this result, we suggest there also be no bigger difference in the performance of voting strategies that were presented, and this may be the point to inventing and testing new strategies by further researchers. Furthermore, adding new variables to a model, just like adding new variables to the X48 variable set, may cause the old variables to lower their importance on the output of models among the Rashomon sets.
Abadi, M., Agarwal, A., Barham, P., Brevdo, E., Chen, Z., Citro, C., et al., et al. (2015).
TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems.
USENIX conference on Operating Systems Design and Implementation.
https://dl.acm.org/doi/10.5555/3026877.3026899
Adadi, A., & Berrada, M. (2018). Peeking inside the black-box: A survey on explainable artificial intelligence (XAI).
IEEE access,
6, 52138–52160.
https://doi.org/10.1109/ACCESS.2018.2870052
Ahmad, M. W., Mourshed, M., & Rezgui, Y. (2017).
Trees vs Neurons: Comparison between random forest and ANN for high-resolution prediction of building energy consumption.
Energy and Buildings,
147, 77--89.
https://doi.org/10.1016/j.enbuild.2017.04.038
Aivodji, U., Arai, H., Fortineau, O., Gambs, S., Hara, S., & Tapp, A. (2019). Fairwashing: The risk of rationalization. In K. Chaudhuri & R. Salakhutdinov (Eds.),
Proceedings of the 36th international conference on machine learning (Vol. 97, pp. 161–170). PMLR.
http://proceedings.mlr.press/v97/aivodji19a.html
Alkahtani, A. S., & Jilani, M. (2019). Predicting return donor and analyzing blood donation time series using data mining techniques.
International Journal of Advanced Computer Science and Applications,
10(8).
https://doi.org/10.14569/IJACSA.2019.0100816
Al-Shawwa, M., Abu-Naser, S., & Nasser, I. (2019). Developing artificial neural network for predicting mobile phone price range (Vol. 3, pp. 1–6).
Andriawan, Z. A., Purnama, S. R., Darmawan, A. S., Ricko, Wibowo, A., Sugiharto, A., & Wijayanto, F. (2020). Prediction of hotel booking cancellation using CRISP-DM. In
2020 4th international conference on informatics and computational sciences (ICICoS) (pp. 1–6).
https://doi.org/10.1109/ICICoS51170.2020.9299011
Anthimopoulos, M., Christodoulidis, S., Ebner, L., Geiser, T., Christe, A., & Mougiakakou, S. (2019). Semantic segmentation of pathological lung tissue with dilated fully convolutional networks.
IEEE Journal of Biomedical and Health Informatics,
23(2), 714–722.
https://doi.org/10.1109/JBHI.2018.2818620
Antonio, N., de Almeida, A., & Nunes, L. (2017). Predicting hotel booking cancellations to decrease uncertainty and increase revenue.
Tourism & Management Studies,
13(2), 25–39.
https://doi.org/10.18089/tms.2017.13203
Antonio, N., de Almeida, A., & Nunes, L. (2019a). An automated machine learning based decision support system to predict hotel booking cancellations.
Data Science Journal,
18(1), 1–20.
https://doi.org/10.5334/dsj-2019-032
Antonio, N., de Almeida, A., & Nunes, L. (2019b). Hotel booking demand datasets.
Data in Brief,
22, 41–49.
https://doi.org/10.1016/j.dib.2018.11.126
Apley, D. (2018).
ALEPlot: Accumulated Local Effects (ALE) Plots and Partial Dependence (PD) Plots.
https://CRAN.R-project.org/package=ALEPlot
Apley, D. W., & Zhu, J. (2020).
Visualizing the effects of predictor variables in black box supervised learning models.
Journal of the Royal Statistical Society Series B,
82(4), 1059–1086.
https://doi.org/10.1111/rssb.12377
Arik, S. O., & Pfister, T. (2021).
TabNet: Attentive Interpretable Tabular Learning.
AAAI Conference on Artificial Intelligence (AAAI).
https://arxiv.org/abs/1908.07442
Arrieta, A. B., Dı́az-Rodrı́guez, N., Del Ser, J., Bennetot, A., Tabik, S., Barbado, A., et al., et al. (2020). Explainable artificial intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI.
Information Fusion,
58, 82–115.
https://doi.org/10.1016/j.inffus.2019.12.012
Arsad, P. M., Buniyamin, N., & Manan, J. A. (2013).
Prediction of engineering students’ academic performance using Artificial Neural Network and Linear Regression: A comparison.
ICEED.
https://doi.org/10.1109/iceed.2013.6908300
Asadi-Aghbolaghi, M., Azad, R., Fathy, M., & Escalera, S. (2020). Multi-level context gating of embedded collective knowledge for medical image segmentation.
https://arxiv.org/abs/2003.05056
Azad, R., Asadi-Aghbolaghi, M., Fathy, M., & Escalera, S. (2019). Bi-directional ConvLSTM u-net with densley connected convolutions. In
2019 IEEE/CVF international conference on computer vision workshop (ICCVW) (pp. 406–415).
https://doi.org/10.1109/ICCVW.2019.00052
Bahel, D., Ghosh, P., Sarkar, A., Lanham, M. A., & Lafayette, W. (2017). Predicting blood donations using machine learning techniques.
http://matthewalanham.com/Students/2017_MWDSI_Final_Bahel.pdf
Baker, M. (2016). Reproducibility crisis. Nature, 533(26), 353–66.
Baniecki, H., Kretowicz, W., Piatyszek, P., Wisniewski, J., & Biecek, P. (2020a).
dalex: Responsible Machine Learning with Interactive Explainability and Fairness in Python.
arXiv:2012.14406.
https://arxiv.org/abs/2012.14406
Baniecki, H., Kretowicz, W., Piatyszek, P., Wisniewski, J., & Biecek, P. (2020b).
dalex: Responsible Machine Learning with Interactive Explainability and Fairness in Python.
arXiv:2012.14406.
https://arxiv.org/abs/2012.14406
Barda, N., Riesel, D., Akriv, A., Levy, J., Finkel, U., Yona, G., et al. (2020).
Developing a COVID-19 mortality risk prediction model when individual-level data are not available.
Nature Communications,
11.
https://doi.org/10.1038/s41467-020-18297-9
Barish, M., Bolourani, S., Lau, L. F., Shah, S., & Zanos, T. P. (2020).
External validation demonstrates limited clinical utility of the interpretable mortality prediction model for patients with COVID-19.
Nature Machine Intelligence,
3, 25–27.
https://doi.org/10.1038/s42256-020-00254-2
Barish, M., Bolourani, S., Lau, L. F., Shah, S., & Zanos, T. P. (2021).
External validation demonstrates limited clinical utility of the interpretable mortality prediction model for patients with COVID-19.
Nature Machine Intelligence,
3(1), 25–27.
https://doi.org/10.1038/s42256-020-00254-2
Barredo Arrieta, A., Díaz-Rodríguez, N., Del Ser, J., Bennetot, A., Tabik, S., Barbado, A., et al. (2020c).
Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI.
Information Fusion,
58, 82–115.
https://doi.org/10.1016/j.inffus.2019.12.012
Barredo Arrieta, A., Díaz-Rodríguez, N., Del Ser, J., Bennetot, A., Tabik, S., Barbado, A., et al. (2020b). Explainable artificial intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI.
Information Fusion,
58, 82–115.
http://www.sciencedirect.com/science/article/pii/S1566253519308103
Barredo Arrieta, A., Díaz-Rodríguez, N., Del Ser, J., Bennetot, A., Tabik, S., Barbado, A., et al. (2020a). Explainable artificial intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI.
Information Fusion,
58, 82–115.
http://www.sciencedirect.com/science/article/pii/S1566253519308103
Barsoum, E., Zhang, C., Ferrer, C. C., & Zhang, Z. (2016). Training deep networks for facial expression recognition with crowd-sourced label distribution. In
Proceedings of the 18th ACM international conference on multimodal interaction (pp. 279–283). New York, NY, USA: Association for Computing Machinery.
https://doi.org/10.1145/2993148.2993165
Belke, A., & Keil, J. (2017). Fundamental determinants of real estate prices: A panel study of german regions, (731). Ruhr Economic Papers.
https://doi.org/10.4419/86788851
Bello-Chavolla, O. Y., Bahena-López, J. P., Antonio-Villa, N. E., Vargas-Vázquez, A., González-Díaz, A., Márquez-Salinas, A., et al. (2020).
Predicting Mortality Due to SARS-CoV-2: A Mechanistic Score Relating Obesity and Diabetes to COVID-19 Outcomes in Mexico.
The Journal of Clinical Endocrinology & Metabolism,
105, 2752--2761.
https://doi.org/10.1210/clinem/dgaa346
Berk, R., Heidari, H., Jabbari, S., Kearns, M., & Roth, A. (2017).
Fairness in Criminal Justice Risk Assessments: The State of the Art.
Sociological Methods & Research.
https://doi.org/10.1177/0049124118782533
Biecek, Przemyslaw. (2018b).
DALEX: Explainers for Complex Predictive Models in R.
Journal of Machine Learning Research,
19(84), 1–5.
http://jmlr.org/papers/v19/18-416.html
Biecek, Przemyslaw. (2018c).
DALEX: Explainers for Complex Predictive Models in R.
Journal of Machine Learning Research,
19(84), 1–5.
https://jmlr.org/papers/v19/18-416.html
Biecek, Przemysław. (2018). DALEX: Explainers for complex predictive models in r. The Journal of Machine Learning Research, 19(1), 3245–3249.
Biecek, Przemyslaw. (2018a).
DALEX: Explainers for Complex Predictive Models in R.
Journal of Machine Learning Research,
19(84), 1–5.
http://jmlr.org/papers/v19/18-416.html
Biecek, Przemyslaw, & Burzykowski, T. (n.d.).
https://ema.drwhy.ai/introduction.html
Biecek, Przemyslaw, & Burzykowski, T. (2021a).
Explanatory Model Analysis. Chapman; Hall/CRC, New York.
https://pbiecek.github.io/ema/
Biecek, Przemyslaw, & Burzykowski, T. (2021b). Explanatory model analysis: Explore, explain, and examine predictive models. CRC Press.
Biecek, Przemyslaw, Maksymiuk, S., & Baniecki, H. (2021).
DALEX: moDel Agnostic Language for Exploration and eXplanation .
https://CRAN.R-project.org/package=DALEX
Bird, S., Dudík, M., Edgar, R., Horn, B., Lutz, R., Milan, V., et al. (2020).
Fairlearn: A toolkit for assessing and improving fairness in AI (No. MSR-TR-2020-32). Microsoft.
https://www.microsoft.com/en-us/research/publication/fairlearn-a-toolkit-for-assessing-and-improving-fairness-in-ai/
Bischl, B., Lang, M., Kotthoff, L., Schiffner, J., Richter, J., Studerus, E., et al. (2016b).
mlr: Machine learning in r.
Journal of Machine Learning Research,
17(170), 1–5.
https://jmlr.org/papers/v17/15-066.html
Bischl, B., Lang, M., Kotthoff, L., Schiffner, J., Richter, J., Studerus, E., et al. (2016a).
mlr: Machine Learning in R.
Journal of Machine Learning Research,
17(170), 1–5.
http://jmlr.org/papers/v17/15-066.html
Bisong, E. (2019). Google colaboratory. In Building machine learning and deep learning models on google cloud platform (pp. 59–64). Springer.
Bradley, A. P. (1997). The use of the area under the ROC curve in the evaluation of machine learning algorithms.
Pattern Recogn.,
30(7), 1145–1159.
https://doi.org/10.1016/S0031-3203(96)00142-2
Breiman, L. (1999). Random forests. UC Berkeley TR567.
Breiman, L. et al. (2001). Statistical modeling: The two cultures (with comments and a rejoinder by the author). Statistical science, 16(3), 199–231.
Browne, M. W. (2000). Cross-validation methods.
Journal of mathematical psychology,
44(1), 108–132.
https://doi.org/10.1006/jmps.1999.1279
Calvert, J., Mao, Q., Rogers, A. J., Barton, C., Jay, M., Desautels, T., et al. (2016). A computational approach to mortality prediction of alcohol use disorder inpatients.
Computers in Biology and Medicine,
75, 74–79. https://doi.org/
https://doi.org/10.1016/j.compbiomed.2016.05.015
Campbell-Kelly, M., Aspray, W., Ensmenger, N., & Yost, J. R. (2018).
Computer: A history of the information machine. Routledge.
https://doi.org/10.4324/9780429495373
Can, A. (1990). The measurement of neighborhood dynamics in urban house prices.
Economic Geography,
66(3), 254–272.
https://doi.org/10.2307/143400
Cao, Y., Liu, X., Xiong, L., & Cai, K. (2020).
Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2: A systematic review and meta-analysis.
Journal of Medical Virology,
92(9), 1449–1459.
https://doi.org/10.1002/jmv.25822
Casadevall, A., & Fang, F. C. (2010). Reproducible science.
Infection and Immunity,
78(12), 4972–4975.
https://doi.org/10.1128/IAI.00908-10
Chen, T., & Guestrin, C. (2016a).
XGBoost: A Scalable Tree Boosting System.
International Conference on Knowledge Discovery and Data Mining (KDD).
https://doi.org/10.1145/2939672.2939785
Chen, T., & Guestrin, C. (2016b).
XGBoost: A Scalable Tree Boosting System.
KDD. https://doi.org/
https://doi.org/10.1145/2939672.2939785
Chollet, F. (2017). Deep learning with python. Manning.
Chouldechova, A. (2016).
Fair Prediction with Disparate Impact: A Study of Bias in Recidivism Prediction Instruments.
Big Data,
5.
https://doi.org/10.1089/big.2016.0047
Chow, J. C. K. (2017, June).
Analysis of Financial Credit Risk Using Machine Learning (Master’s thesis). Aston University. Retrieved from
https://www.researchgate.net/publication/318959365_Analysis_of_Financial_Credit_Risk_Using_Machine_Learning
Chowdhury, M. E. H., Rahman, T., Khandakar, A., Mazhar, R., Kadir, M. A., Mahbub, Z. B., et al. (2020). Can AI help in screening viral and COVID-19 pneumonia?
IEEE Access,
8, 132665–132676.
https://doi.org/10.1109/ACCESS.2020.3010287
Christe, A. A. M. D., Andreas MD∗; Peters. (2019). Computer-aided diagnosis of pulmonary fibrosis using deep learning and CT images.
Investigative Radiology,
54, 627–632.
https://doi.org/10.1097/RLI.0000000000000574
Chu, X., Ilyas, I. F., Krishnan, S., & Wang, J. (2016). Data cleaning: Overview and emerging challenges. In Proceedings of the 2016 international conference on management of data (pp. 2201–2206).
Clark, K., Vendt, B., Smith, K., Freymann, J., Kirby, J., Koppel, P., et al. (2013). The cancer imaging archive (TCIA): Maintaining and operating a public information repository.
Journal of Digital Imaging,
26(6), 1045–1057.
https://doi.org/10.1007/s10278-013-9622-7
Code of Federal Regulations. (1978). SECTION 4D, UNIFORM GUIDELINES ON EMPLOYEE SELECTION PROCEDURES (1978).
https://www.govinfo.gov/content/pkg/CFR-2014-title29-vol4/xml/CFR-2014-title29-vol4-part1607.xml
Cohen, J. P., Morrison, P., Dao, L., Roth, K., Duong, T. Q., & Ghassemi, M. (2020a). COVID-19 image data collection: Prospective predictions are the future.
arXiv 2006.11988.
https://github.com/ieee8023/covid-chestxray-dataset
Cohen, J. P., Morrison, P., Dao, L., Roth, K., Duong, T. Q., & Ghassemi, M. (2020b). COVID-19 image data collection: Prospective predictions are the future.
arXiv 2006.11988.
https://github.com/ieee8023/covid-chestxray-dataset
Computing Machinery, A. for. (2018). Artifact review and badging.
https://www.acm.org/publications/policies/artifact-review badging
Conway, J. (2018, January).
Artificial Intelligence and Machine Learning : Current Applications in Real Estate (PhD thesis). Retrieved from
https://dspace.mit.edu/bitstream/handle/1721.1/120609/1088413444-MIT.pdf
Corbett-Davies, S., Pierson, E., Feller, A., Goel, S., & Huq, A. (2017).
Algorithmic Decision Making and the Cost of Fairness.
https://doi.org/10.1145/3097983.3098095
Culp, M., Johnson, K., & Michailidis, G. (2016).
Ada: The r package ada for stochastic boosting.
https://CRAN.R-project.org/package=ada
Darwiche, M., Feuilloy, M., Bousaleh, G., & Schang, D. (2010). Prediction of blood transfusion donation, 51–56.
https://doi.org/10.1109/RCIS.2010.5507363
Das, S., Cashman, D., Chang, R., & Endert, A. (2019). BEAMES: Interactive multimodel steering, selection, and inspection for regression tasks.
IEEE Computer Graphics and Applications,
39(5), 20–32.
https://doi.org/10.1109/MCG.2019.2922592
Davis, J., & Goadrich, M. (2006). The relationship between precision-recall and ROC curves. In
Proceedings of the 23rd international conference on machine learning (pp. 233–240). New York, NY, USA: Association for Computing Machinery.
https://doi.org/10.1145/1143844.1143874
Deo, R. C. (2015). Machine learning in medicine.
Circulation,
132(20), 1920–1930.
https://doi.org/10.1161/CIRCULATIONAHA.115.001593
Desai, B., S. (2020). Data from chest imaging with clinical and genomic correlates representing a rural COVID-19 positive population [data set].
The Cancer Imaging Archive.
https://doi.org/10.7937/tcia.2020.py71-5978
Detrano, R., Janosi, A., Steinbrunn, W., Pfisterer, M., Schmid, J.-J., Sandhu, S., et al. (1989). International application of a new probability algorithm for the diagnosis of coronary artery disease.
The American Journal of Cardiology,
64(5), 304–310.
https://doi.org/10.1016/0002-9149(89)90524-9
Detrano, R., Yiannikas, J., Salcedo, E. E., Rincon, G., Go, R. T., Williams, G., & Leatherman, J. (1984). Bayesian probability analysis: A prospective demonstration of its clinical utility in diagnosing coronary disease.
Circulation,
69(3), 541—547.
https://doi.org/10.1161/01.CIR.69.3.541
Dong, J., & Rudin, C. (2020). Exploring the cloud of variable importance for the set of all good models. Nature Machine Intelligence, 2(12), 810–824.
Du, X., Cai, Y., Wang, S., & Zhang, L. (2016). Overview of deep learning. In 2016 31st youth academic annual conference of chinese association of automation (YAC) (pp. 159–164). IEEE.
Dua, D., & Graff, C. (2017).
UCI machine learning repository.
http://archive.ics.uci.edu/ml
Dubin, R. A. (1998). Predicting house prices using multiple listings data.
The Journal of Real Estate Finance and Economics.
https://doi.org/10.1023/A:1007751112669
Dupuis, C., De Montmollin, E., Neuville, M., Mourvillier, B., Ruckly, S., & Timsit, J. F. (2021).
Limited applicability of a COVID-19 specific mortality prediction rule to the intensive care setting.
Nature Machine Intelligence,
3(1), 20–22.
https://doi.org/10.1038/s42256-020-00252-4
Dwork, C., Hardt, M., Pitassi, T., Reingold, O., & Zemel, R. (2012). Fairness through awareness.
ITCS.
https://doi.org/10.1145/2090236.2090255
Falk, M., & Vieru, M. (2018). Modelling the cancellation behaviour of hotel guests.
International Journal of Contemporary Hospitality Management,
30(10), 3100–3116.
https://doi.org/10.1108/ijchm-08-2017-0509
Fan, C., Cui, Z., & Zhong, X. (2018). House prices prediction with machine learning algorithms. In
Proceedings of the 2018 10th international conference on machine learning and computing (pp. 6–10). New York, NY, USA: Association for Computing Machinery.
https://doi.org/10.1145/3195106.3195133
Fawcett, T. (2006). An introduction to ROC analysis.
Pattern Recognition Letters,
27(8), 861–874. https://doi.org/
https://doi.org/10.1016/j.patrec.2005.10.010
Finlayson, S. G., Chung, H. W., Kohane, I. S., & Beam, A. L. (2018). Adversarial attacks against medical deep learning systems. arXiv preprint arXiv:1804.05296.
Fisher, A., Rudin, C., & Dominici, F. (2018b).
All Models are Wrong, but Many are Useful: Learning a Variable’s Importance by Studying an Entire Class of Prediction Models Simultaneously.
arXiv.
https://arxiv.org/abs/1801.01489
Fisher, A., Rudin, C., & Dominici, F. (2018a).
All Models are Wrong, but Many are Useful: Learning a Variable’s Importance by Studying an Entire Class of Prediction Models Simultaneously.
arXiv.
https://arxiv.org/abs/1801.01489
Fisher, A., Rudin, C., & Dominici, F. (2019a). All models are wrong, but many are useful: Learning a variable’s importance by studying an entire class of prediction models simultaneously.
Journal of Machine Learning Research,
20(177), 1–81.
http://jmlr.org/papers/v20/18-760.html
Fisher, A., Rudin, C., & Dominici, F. (2019d).
All Models are Wrong, but Many are Useful: Learning a Variable’s Importance by Studying an Entire Class of Prediction Models Simultaneously.
Journal of Machine Learning Research,
20(177), 1–81.
http://jmlr.org/papers/v20/18-760.html
Fisher, A., Rudin, C., & Dominici, F. (2019e). All models are wrong, but many are useful: Learning a variable’s importance by studying an entire class of prediction models simultaneously. Journal of Machine Learning Research, 20(177), 1–81.
Fisher, A., Rudin, C., & Dominici, F. (2019b). All models are wrong, but many are useful: Learning a variable’s importance by studying an entire class of prediction models simultaneously.
Journal of Machine Learning Research,
20(177), 1–81.
http://jmlr.org/papers/v20/18-760.html
Fisher, A., Rudin, C., & Dominici, F. (2019c). All models are wrong, but many are useful: Learning a variable’s importance by studying an entire class of prediction models simultaneously. Journal of Machine Learning Research, 20(177), 1–81.
Fisher, R. A. (1922). On the interpretation of χ2 from contingency tables, and the calculation of p.
Journal of the Royal Statistical Society,
85(1), 87–94.
http://www.jstor.org/stable/2340521
Friedman, J. H. (2000a). Greedy function approximation: A gradient boosting machine. Annals of Statistics, 29, 1189–1232.
Friedman, J. H. (2000b).
Greedy Function Approximation: A Gradient Boosting Machine.
Annals of Statistics,
29, 1189–1232.
https://doi.org/10.1214/aos/1013203451
Friedman, J. H. (2000c).
Greedy Function Approximation: A Gradient Boosting Machine.
Annals of Statistics,
29, 1189–1232.
https://doi.org/10.1214/aos/1013203451
Ge, X., Runeson, G., & Lam, K. C. (2021). Forecasting hong kong housing prices: An artificial neural network approach.
Genders, T. S. S., Steyerberg, E. W., Alkadhi, H., Leschka, S., Desbiolles, L., Nieman, K., et al. (2011). A clinical prediction rule for the diagnosis of coronary artery disease: Validation, updating, and extension.
European Heart Journal,
32(11), 1316–1330.
https://doi.org/10.1093/eurheartj/ehr014
Géron, A. (2017). Hands-on machine learning with scikit-learn and TensorFlow : Concepts, tools, and techniques to build intelligent systems. O’Reilly Media.
Ghysels, E., Plazzi, A., Valkanov, R., & Torous, W. (2013). Chapter 9 - forecasting real estate prices,
2, 509–580. https://doi.org/
https://doi.org/10.1016/B978-0-444-53683-9.00009-8
Gilpin, L. H., Bau, D., Yuan, B. Z., Bajwa, A., Specter, M., & Kagal, L. (2018). Explaining explanations: An overview of interpretability of machine learning. In
2018 IEEE 5th international conference on data science and advanced analytics (DSAA) (pp. 80–89). IEEE.
https://doi.org/10.1109/DSAA.2018.00018
Glauner, P. (2021). An assessment of the AI regulation proposed by the european commission.
https://arxiv.org/abs/2105.15133
GOLDNER, M. C., ZAMORA, M. C., DI LEO LIRA, P., GIANNINOTO, H., & BANDONI, A. (2009). EFFECT OF ETHANOL LEVEL IN THE PERCEPTION OF AROMA ATTRIBUTES AND THE DETECTION OF VOLATILE COMPOUNDS IN RED WINE. Journal of sensory studies, 24(2), 243–257.
Goldstein, A., Kapelner, A., Bleich, J., & Pitkin, E. (2014). Peeking inside the black box: Visualizing statistical learning with plots of individual conditional expectation.
Goldstein, A., Kapelner, A., Bleich, J., & Pitkin, E. (2015).
Peeking Inside the Black Box: Visualizing Statistical Learning With Plots of Individual Conditional Expectation.
Journal of Computational and Graphical Statistics,
24(1), 44–65.
https://doi.org/10.1080/10618600.2014.907095
Goodman, B., & Flaxman, S. (2017). European union regulations on algorithmic decision-making and a
“right to explanation.” AI Magazine,
38(3), 50–57.
https://doi.org/10.1609/aimag.v38i3.2741
Gosiewska, A., & Biecek, P. (2019b).
Do Not Trust Additive Explanations.
arXiv.
https://arxiv.org/abs/1903.11420v3
Gosiewska, A., & Biecek, P. (2019a). Do not trust additive explanations.
arXiv preprint arXiv:1903.11420.
https://doi.org/arXiv:1903.11420
Gosiewska, A., & Biecek, P. (2020). Do not trust additive explanations.
https://arxiv.org/abs/1903.11420
Goyal, S. (2020, November). Credit card customers.
Kaggle.
https://www.kaggle.com/sakshigoyal7/credit-card-customers
Greenwell, Brandon M. (2017c).
pdp: An R Package for Constructing Partial Dependence Plots.
The R Journal,
9(1), 421–436.
http://doi.org/10.32614/RJ-2017-016
Greenwell, Brandon M. (2017a). Pdp: An r package for constructing partial dependence plots.
R J.,
9(1), 421.
https://doi.org/10.32614/RJ-2017-016
Greenwell, Brandon M. (2017b).
pdp: An R Package for Constructing Partial Dependence Plots.
The R Journal,
9(1), 421–436.
http://doi.org/10.32614/RJ-2017-016
Greenwell, B., Boehmke, B., Cunningham, J., & Developers, G. (2020).
Gbm: Generalized boosted regression models.
https://CRAN.R-project.org/package=gbm
Gunning, D., Stefik, M., Choi, J., Miller, T., Stumpf, S., & Yang, G.-Z. (2019). XAI—explainable artificial intelligence.
Science Robotics,
4(37).
https://doi.org/10.1126/scirobotics.aay7120
Hanley, J. A. (2014).
Receiver Operating Characteristic (ROC) Curves.
Wiley StatsRef: Statistics Reference Online.
https://doi.org/10.1002/9781118445112.stat05255
Hardt, M., Price, E., Price, E., & Srebro, N. (2016).
Equality of Opportunity in Supervised Learning.
NeurIPS.
https://papers.nips.cc/paper/2016/hash/9d2682367c3935defcb1f9e247a97c0d-Abstract.html
Heyman, A., & Sommervoll, D. (2019). House prices and relative location.
Cities,
95, 102373.
https://doi.org/10.1016/j.cities.2019.06.004
Holzinger, A. (2016). Interactive machine learning for health informatics: When do we need the human-in-the-loop?
Brain Informatics,
3, 119–131.
https://doi.org/10.1007/s40708-016-0042-6
Holzinger, A. (2021). Explainable AI and multi-modal causability in medicine.
i-com,
19(3), 171–179.
https://doi.org/10.1515/icom-2020-0024
Holzinger, A., Biemann, C., Pattichis, C., & Kell, D. (2017). What do we need to build explainable AI systems for the medical domain?
Holzinger, A., Langs, G., Denk, H., Zatloukal, K., & Müller, H. (2019). Causability and explainabilty of artificial intelligence in medicine.
Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery,
9.
https://doi.org/10.1002/widm.131
Johnson, A. E. W., Pollard, T. J., & Mark, R. G. (2017). Reproducibility in critical care: A mortality prediction case study. In F. Doshi-Velez, J. Fackler, D. Kale, R. Ranganath, B. Wallace, & J. Wiens (Eds.),
Proceedings of the 2nd machine learning for healthcare conference (Vol. 68, pp. 361–376). Boston, Massachusetts: PMLR.
http://proceedings.mlr.press/v68/johnson17a.html
Johnson, A. E., Pollard, T. J., Shen, L., Li-Wei, H. L., Feng, M., Ghassemi, M., et al. (2016). MIMIC-III, a freely accessible critical care database. Scientific data, 3(1), 1–9.
Jordão, A. M., Vilela, A., & Cosme, F. (2015). From sugar of grape to alcohol of wine: Sensorial impact of alcohol in wine.
Beverages,
1(4), 292–310.
https://doi.org/10.3390/beverages1040292
Kaladharan, S., Vishvanathan, S., Gopalakrishnan, E. A., & Kp, S. (2020). Explainable artificial intelligence for heart rate variability in ECG signal.
Healthcare Technology Letters,
7, 146–154.
https://doi.org/10.1049/htl.2020.0033
Karim, M. R., Döhmen, T., Rebholz-Schuhmann, D., Decker, S., Cochez, M., & Beyan, O. (2020). DeepCOVIDExplainer: Explainable COVID-19 diagnosis from chest x-ray images. IEEE.
https://doi.org/10.1109/BIBM49941.2020.9313304
Kather, J. N., Zöllner, F. G., Bianconi, F., Melchers, S. M., Schad, L. R., Gaiser, T., et al. (2016, May).
Collection of textures in colorectal cancer histology. Zenodo.
https://doi.org/10.5281/zenodo.53169
Kaushal, A., Altman, R., & Langlotz, C. (2020).
Health Care AI Systems Are Biased.
Scientific American.
https://www.scientificamerican.com/article/health-care-ai-systems-are-biased
Kennedy, K. (2013).
Credit scoring using machine learning (PhD thesis). Technological University Dublin. Retrieved from
https://arrow.tudublin.ie/sciendoc/137/
Khalifa, N. E. M., Taha, M. H. N., Hassanien, A. E., & Elghamrawy, S. (2020). Detection of coronavirus (COVID-19) associated pneumonia based on generative adversarial networks and a fine-tuned deep transfer learning model using chest x-ray dataset.
https://arxiv.org/abs/2004.01184
Khedkar, S., Subramanian, V., Shinde, G., & Gandhi, P. (2019). Explainable AI in healthcare.
SSRN Electronic Journal.
https://doi.org/10.2139/ssrn.3367686
Kieseberg, P., Schantl, J., Fruehwirt, P., Weippl, E., & Holzinger, A. (2015). Witnesses for the doctor in the loop,
9250, 369–378.
https://doi.org/10.1007/978-3-319-23344-4_36
Klambauer, G., Unterthiner, T., Mayr, A., & Hochreiter, S. (2017).
Self-Normalizing Neural Networks.
arXiv:1706.02515.
https://arxiv.org/abs/1706.02515
Komisarczyk, Konrad and Maksymiuk, Szymon and Koźmiński, Paweł and Biecek, Przemysław. (2020).
treeshap: Fast SHAP values computation for ensemble models.
R package.
https://github.com/ModelOriented/treeshap
Kowsari, K., Brown, D. E., Heidarysafa, M., Jafari Meimandi, K., Gerber, M. S. and, & Barnes, L. E. (2017). HDLTex: Hierarchical deep learning for text classification. In Machine learning and applications (ICMLA), 2017 16th IEEE international conference on. IEEE.
Kowsari, K., Heidarysafa, M., Brown, D. E., Meimandi, K. J., & Barnes, L. E. (2018b). Rmdl: Random multimodel deep learning for classification. In Proceedings of the 2nd international conference on information system and data mining (pp. 19–28).
Kowsari, K., Heidarysafa, M., Brown, D. E., Meimandi, K. J., & Barnes, L. E. (2018a). Rmdl: Random multimodel deep learning for classification. In Proceedings of the 2nd international conference on information system and data mining (pp. 19–28).
Krittanawong, C., Zhang, H., Wang, Z., Aydar, M., & Kitai, T. (2017). Artificial intelligence in precision cardiovascular medicine.
Journal of the American College of Cardiology,
69(21), 2657–2664.
https://doi.org/10.1016/j.jacc.2017.03.571
Law, S. (2017). Defining street-based local area and measuring its effect on house price using a hedonic price approach: The case study of metropolitan london.
Cities,
60, 166–179.
https://doi.org/10.1016/j.cities.2016.08.008
Lerman, R. I., & Yitzhaki, S. (1984). A note on the calculation and interpretation of the gini index.
Economics Letters,
15(3-4), 363–368.
https://doi.org/10.1016/0165-1765(84)90126-5
Li, X., Ge, P., Zhu, J., Li, H., Graham, J., Singer, A., et al. (2020).
Deep learning prediction of likelihood of ICU admission and mortality in COVID-19 patients using clinical variables.
PeerJ,
8.
https://peerj.com/articles/10337/
Liaw, A., & Wiener, M. (2002a). Classification and regression by randomForest.
R News,
2(3), 18–22.
https://CRAN.R-project.org/doc/Rnews/
Liaw, A., & Wiener, M. (2002b). Classification and regression by randomForest.
R News,
2(3), 18–22.
https://CRAN.R-project.org/doc/Rnews/
Linardatos, P., Papastefanopoulos, V., & Kotsiantis, S. (2021).
Explainable AI: A Review of Machine Learning Interpretability Methods.
Entropy.
https://www.mdpi.com/1099-4300/23/1/18/pdf
Liu, C., Gao, C., Xia, X., Lo, D., Grundy, J., & Yang, X. (2020). On the replicability and reproducibility of deep learning in software engineering.
https://arxiv.org/abs/2006.14244
Loyola-González, O. (2019). Black-box vs. White-box: Understanding their advantages and weaknesses from a practical point of view.
IEEE Access,
7, 154096–154113.
https://doi.org/10.1109/ACCESS.2019.2949286
Łukasz Rączkowski, J. Z., Marcin Możejko. (2019). ARA: Accurate, reliable and active histopathological image classification framework with bayesian deep learning.
Springer Nature,
14, 1–11.
https://doi.org/10.1038/s41598-019-50587-1
Lundberg, Scott M., Erion, G. G., & Lee, S.-I. (2019).
Consistent Individualized Feature Attribution for Tree Ensembles.
ICML Workshop.
https://arxiv.org/abs/1802.03888
Lundberg, Scott M., & Lee, S.-I. (2017). A unified approach to interpreting model predictions. In I. Guyon, U. V. Luxburg, S. Bengio, H. Wallach, R. Fergus, S. Vishwanathan, & R. Garnett (Eds.),
Advances in neural information processing systems 30 (pp. 4765–4774). Montreal: Curran Associates.
http://papers.nips.cc/paper/7062-a-unified-approach-to-interpreting-model-predictions.pdf
Lundberg, S., & Lee, S.-I. (2017). A unified approach to interpreting model predictions.
arXiv preprint arXiv:1705.07874.
https://doi.org/arXiv:1705.07874
Lundegård, Z. (2019).
Current AI technologies for medical imaging and ethical dilemmas created by them (Master’s thesis). Åbo Akademi. Retrieved from
https://core.ac.uk/download/pdf/186507687.pdf
M. Barhoom, A., Abu-Naser, S., Abu-Nasser, B., Alajrami, E., Musleh, M., & Khalil, A. (2019). Blood donation prediction using artificial neural network, 1–7.
https://philarchive.org/archive/BARBDP-14
Ma, X., Ng, M., Xu, S., Xu, Z., Qiu, H., Liu, Y., et al. (2020).
Development and validation of prognosis model of mortality risk in patients with COVID-19.
Epidemiology and Infection,
148.
http://doi.org/10.1017/S0950268820001727
Maksymiuk, S., & Biecek, P. (2020b).
DALEXtra: Extension for ’DALEX’ Package.
https://CRAN.R-project.org/package=DALEXtra
Maksymiuk, S., & Biecek, P. (2020a).
DALEXtra: Extension for ’DALEX’ Package.
https://CRAN.R-project.org/package=DALEXtra
Maksymiuk, S., Gosiewska, A., & Biecek, P. (2020b). Landscape of r packages for eXplainable artificial intelligence.
arXiv.
https://arxiv.org/abs/2009.13248
Maksymiuk, S., Gosiewska, A., & Biecek, P. (2020a). Landscape of r packages for eXplainable artificial intelligence.
arXiv preprint arXiv:2009.13248.
https://doi.org/arXiv:2009.13248
Maksymiuk, S., Gosiewska, A., & Biecek, P. (2021). Landscape of r packages for eXplainable artificial intelligence.
https://arxiv.org/abs/2009.13248
Mattern, F., Staake, T., & Weiss, M. (2010). ICT for green: How computers can help us to conserve energy. In
Proceedings of the 1st international conference on energy-efficient computing and networking (pp. 1–10).
https://doi.org/10.1145/1791314.1791316
Mendez, D., Graziotin, D., Wagner, S., & Seibold, H. (2020). Open science in software engineering.
Contemporary Empirical Methods in Software Engineering, 477–501.
https://doi.org/10.1007/978-3-030-32489-6_17
Meyer, D., Dimitriadou, E., Hornik, K., Weingessel, A., & Leisch, F. (2020).
e1071: Misc functions of the department of statistics, probability theory group (formerly: E1071), TU wien.
https://CRAN.R-project.org/package=e1071
Meyer, D., Dimitriadou, E., Hornik, K., Weingessel, A., Leisch, F., Chang, C.-C., & Lin, C.-C. (2021).
e1071: Misc Functions of the Department of Statistics, Probability Theory Group.
R package.
https://CRAN.R-project.org/package=e1071
Mikolov, T., Sutskever, I., Chen, K., Corrado, G., & Dean, J. (2013). Distributed representations of words and phrases and their compositionality.
CoRR,
abs/1310.4546.
http://arxiv.org/abs/1310.4546
Molnar, C. (2019).
Interpretable Machine Learning: A Guide for Making Black Box Models Explainable.
https://christophm.github.io/interpretable-ml-book
Neff, T., Payer, C., Stern, D., & Urschler, M. (2017). Generative adversarial network based synthesis for supervised medical image segmentation. In Proc. OAGM and ARW joint workshop.
Ngoc Anh, H. (2016). Smartphone industry: The new era of competition and strategy (pp. 1–46).
Oktay, O., Schlemper, J., Folgoc, L. L., Lee, M., Heinrich, M., Misawa, K., et al., et al. (2018). Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:1804.03999.
Pace, R. K., & Barry, R. (1997). Sparse spatial autoregressions. Statistics & Probability Letters, 33(3), 291–297.
Pal, M. (2005). Random forest classifier for remote sensing classification.
International journal of remote sensing,
26(1), 217–222.
https://doi.org/10.1080/01431160412331269698
Pandala, S. R. (2019).
Lazy Predict.
Python package.
https://github.com/shankarpandala/lazypredict
Park, B., & Bae, J. (2015).
Using machine learning algorithms for housing price prediction: The case of Fairfax County, Virginia housing data.
Expert Systems with Applications,
42.
https://doi.org/10.1016/j.eswa.2014.11.040
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., et al. (2011a). Scikit-learn: Machine learning in Python. Journal of Machine Learning Research, 12, 2825–2830.
Pedregosa, Fabian, Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P., Weiss, R., Dubourg, V., Vanderplas, J., Passos, A., Cournapeau, D., Brucher, M., Perrot, M., & Duchesnay, Édouard. (2011b).
Scikit-learn: Machine Learning in Python.
Journal of Machine Learning Research,
12(85), 2825–2830.
http://jmlr.org/papers/v12/pedregosa11a.html
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., et al. (2011b). Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research, 12, 2825–2830.
Pedregosa, Fabian, Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P., Weiss, R., Dubourg, V., Vanderplas, J., Passos, A., Cournapeau, D., Brucher, M., Perrot, M., & Duchesnay, douard. (2011a). Scikit-learn: Machine learning in Python. Journal of Machine Learning Research, 12, 2825–2830.
Pekala, K., Woznica, K., & Biecek, P. (2021a). Triplot: Model agnostic measures and visualisations for variable importance in predictive models that take into account the hierarchical correlation structure.
CoRR,
abs/2104.03403.
https://arxiv.org/abs/2104.03403
Pekala, K., Woznica, K., & Biecek, P. (2021b). Triplot: Model agnostic measures and visualisations for variable importance in predictive models that take into account the hierarchical correlation structure.
arXiv preprint arXiv:2104.03403.
https://doi.org/arXiv:2104.03403
Peng, Z., Huang, Q., & Han, Y. (2019). Model research on forecast of second-hand house price in chengdu based on XGboost algorithm, 168–172.
https://doi.org/10.1109/ICAIT.2019.8935894
Pineau, J., Vincent-Lamarre, P., Sinha, K., Larivière, V., Beygelzimer, A., d’Alché-Buc, F., et al. (2020). Improving reproducibility in machine learning research
(A report from the NeurIPS 2019 reproducibility program).
CoRR,
abs/2003.12206.
https://arxiv.org/abs/2003.12206
Plumb, G., Molitor, D., & Talwalkar, A. (2019). Model agnostic supervised local explanations.
https://arxiv.org/abs/1807.02910
Probst, P., Boulesteix, A.-L., & Bischl, B. (2019). Tunability: Importance of hyperparameters of machine learning algorithms.
Journal of Machine Learning Research,
20(53), 1–32.
http://jmlr.org/papers/v20/18-444.html
Quandt, R. E. (1983). Computational problems and methods.
Handbook of econometrics,
1, 699–764.
https://doi.org/10.1016/S1573-4412(83)01016-8
Quanjel, M. J. R., Holten, T. C. van, Gunst-van der Vliet, P. C., Wielaard, J., Karakaya, B., Söhne, M., et al. (2021). Replication of a mortality prediction model in dutch patients with COVID-19.
Nature Machine Intelligence,
3(1), 23–24.
https://doi.org/10.1038/s42256-020-00253-3
Quanjel, M. J. R., Holten, T. C. van, Vliet, P. C. G. der, Wielaard, J., Karakaya, B., Söhne, M., et al. (2021).
Replication of a mortality prediction model in Dutch patients with COVID-19.
Nature Machine Intelligence,
3, 23–24.
https://doi.org/10.1038/s42256-020-00253-3
R Core Team. (2018).
R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
https://www.R-project.org/
R Core Team. (2021).
R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
https://www.R-project.org/
Raghavan, V., Bollmann, P., & Jung, G. S. (1989). A critical investigation of recall and precision as measures of retrieval system performance.
ACM Trans. Inf. Syst.,
7(3), 205–229.
https://doi.org/10.1145/65943.65945
Rahimzadeh, M., Attar, A., & Sakhaei, S. M. (2021). A fully automated deep learning-based network for detecting COVID-19 from a new and large lung CT scan dataset.
Biomedical Signal Processing and Control, 102588. https://doi.org/
https://doi.org/10.1016/j.bspc.2021.102588
Rahman, T., Khandakar, A., Qiblawey, Y., Tahir, A., Kiranyaz, S., Abul Kashem, S. B., et al. (2021). Exploring the effect of image enhancement techniques on COVID-19 detection using chest x-ray images.
Computers in Biology and Medicine,
132, 104319. https://doi.org/
https://doi.org/10.1016/j.compbiomed.2021.104319
Rai, A. (2020).
Explainable AI: from black box to glass box.
Journal of the Academy of Marketing Science,
48, 137–141.
https://link.springer.com/article/10.1007/s11747-019-00710-5
Religia, Y., Pranoto, G. T., & Santosa, E. D. (2020). South german credit data classification using random forest algorithm to predict bank credit receipts.
JISA (Jurnal Informatika dan Sains),
3(2), 62–66.
https://doi.org/10.31326/jisa.v3i2.837
Riasi, A., Schwartz, Z., & Chen, C.-C. (2019). A paradigm shift in revenue management? The new landscape of hotel cancellation policies.
Journal of Revenue and Pricing Management,
18(6), 434–440.
https://doi.org/10.1057/s41272-019-00189-3
Ribeiro, M. T., Singh, S., & Guestrin, C. (2016a). "Why should I trust you?": Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, KDD san francisco, CA (pp. 1135–1144). New York, NY: Association for Computing Machinery.
Ribeiro, M. T., Singh, S., & Guestrin, C. (2016b).
"Why Should I Trust You?": Explaining the Predictions of Any Classifier. In
Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, san francisco, CA, USA, august 13-17, 2016 (pp. 1135–1144).
https://doi.org/10.18653/v1/n16-3020
Ribeiro, M. T., Singh, S., & Guestrin, C. (2016c). " why should i trust you?" Explaining the predictions of any classifier. In
Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining (pp. 1135–1144).
https://doi.org/10.1145/2939672.2939778
Roberto Castro Sundin, A. S. G. &. S. W., Tony Rönnqvist. (2020). Siamesifying the COVID-net.
https://people.kth.se/~rosun/deep-learning/
Ruder, S. (2017). An overview of multi-task learning in deep neural networks.
https://arxiv.org/abs/1706.05098
Rudin, C. (2019b). Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead.
Nature Machine Intelligence,
1(5), 206–215.
https://doi.org/10.1038/s42256-019-0048-x
Rudin, C. (2019a). Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead.
Nature Machine Intelligence,
1, 206–215.
https://doi.org/10.1038/s42256-019-0048
Rudin, C., Chen, C., Chen, Z., Huang, H., Semenova, L., & Zhong, C. (2021). Interpretable machine learning: Fundamental principles and 10 grand challenges. arXiv preprint arXiv:2103.11251.
Saarela, M., & Jauhiainen, S. (2021). Comparison of feature importance measures as explanations for classification models.
SN Applied Sciences,
3(2).
https://doi.org/10.1007/s42452-021-04148-9
Saito, T., & Rehmsmeier, M. (2015). The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets.
Plos One,
10(3).
https://doi.org/10.1371/journal.pone.0118432
Sánchez-Medina, A. J., & C-Sánchez, E. (2020). Using machine learning and big data for efficient forecasting of hotel booking cancellations.
International Journal of Hospitality Management,
89, 102546.
https://doi.org/10.1016/j.ijhm.2020.102546
Sandfort, V., Yan, K., Pickhardt, P. J., & Summers, R. M. (2019). Data augmentation using generative adversarial networks (CycleGAN) to improve generalizability in CT segmentation tasks. Scientific reports, 9(1), 1–9.
Semenova, L., Rudin, C., & Parr, R. (2019). A study in rashomon curves and volumes: A new perspective on generalization and model simplicity in machine learning. arXiv preprint arXiv:1908.01755.
Shea, A. M., Hammill, B. G., Curtis, L. H., Szczech, L. A., & Schulman, K. A. (2008). Medical costs of abnormal serum sodium levels.
Journal of the American Society of Nephrology,
19(4), 764–770.
https://doi.org/10.1681/ASN.2007070752
Siler, W. (2013). Computers in life science research (Vol. 2). Springer Science & Business Media.
Singh, R. (2021). Exploratory data analysis and customer segmentation for smartphones.
Slack, D., Hilgard, S., Jia, E., Singh, S., & Lakkaraju, H. (2020). Fooling LIME and SHAP: Adversarial attacks on post hoc explanation methods. In
Proceedings of the AAAI/ACM conference on AI, ethics, and society (pp. 180–186). New York, NY, USA: Association for Computing Machinery.
https://doi.org/10.1145/3375627.3375830
Smith, S. J., Parsa, H. G., Bujisic, M., & van der Rest, J.-P. (2015). Hotel cancelation policies, distributive and procedural fairness, and consumer patronage: A study of the lodging industry.
Journal of Travel & Tourism Marketing,
32, 886–906.
https://doi.org/10.1080/10548408.2015.1063864
Sofaer, H. R., Hoeting, J. A., & Jarnevich, C. S. (2019).
The area under the precision-recall curve as a performance metric for rare binary events.
Methods in Ecology and Evolution,
10(4), 565–577.
https://doi.org/10.1111/2041-210X.13140
Staniak, M., & Biecek, P. (2018a). Explanations of model predictions with live and breakDown packages.
Staniak, M., & Biecek, P. (2018b). Explanations of model predictions with live and breakDown packages.
arXiv preprint arXiv:1804.01955.
https://doi.org/10.32614/RJ-2018-072
Staniak, M., Kuzba, M., & Biecek, P. (2018).
Local explanations of complex machine learning models.
https://doi.org/10.13140/RG.2.2.23637.58084
Suzuki, K. (2017). Overview of deep learning in medical imaging. Radiological physics and technology, 10(3), 257–273.
Tang, F., Xiao, C., Wang, F., & Zhou, J. (2018). Predictive modeling in urgent care: A comparative study of machine learning approaches. Jamia Open, 1(1), 87–98.
Tatman, R., VanderPlas, J., & Dane, S. (2018). A practical taxonomy of reproducibility for machine learning research.
Thompson, N. C., Greenewald, K., Lee, K., & Manso, G. F. (2020). The computational limits of deep learning. arXiv preprint arXiv:2007.05558.
Tonekaboni, S., Joshi, S., McCradden, M. D., & Goldenberg, A. (2019).
What Clinicians Want: Contextualizing Explainable Machine Learning for Clinical End Use.
Machine Learning for Healthcare.
http://proceedings.mlr.press/v106/tonekaboni19a.html
Tsai, Simpson, E. (2020). Data from the medical imaging data resource center - RSNA international COVID radiology database release 1a - chest CT covid+ (MIDRC-RICORD-1a).
The Cancer Imaging Archive.
https://doi.org/10.7937/VTW4-X588
Tsai, Simpson, E. B. (2021). Medical imaging data resource center (MIDRC) - RSNA international COVID open research database (RICORD) release 1b - chest CT covid- [data set]. The Cancer Imaging Archive.
Turney, P. D. (1995). Cost-sensitive classification: Empirical evaluationof a hybrid genetic decision tree induction algorithm.
https://www.jair.org/index.php/jair/article/view/10129/23991
Ucar, F., & Korkmaz, D. (2020). COVIDiagnosis-net: Deep bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from x-ray images. Medical Hypotheses, 140, 109761–109761.
van Buuren, S., & Groothuis-Oudshoorn, K. (2011).
mice: Multivariate imputation by chained equations in r.
Journal of Statistical Software,
45(3), 1–67.
https://www.jstatsoft.org/v45/i03/
Vandewalle, P., Kovacevic, J., & Vetterli, M. (2009). Reproducible research in signal processing. IEEE Signal Processing Magazine, 26(3), 37–47.
Vanschoren, J., Rijn, J. N. van, Bischl, B., & Torgo, L. (2013). OpenML: Networked science in machine learning.
SIGKDD Explorations,
15(2), 49–60.
https://doi.org/10.1145/2641190.2641198
Varma, A., Sarma, A., Doshi, S., & Nair, R. (2018). House price prediction using machine learning and neural networks, 1936–1939.
https://doi.org/10.1109/ICICCT.2018.8473231
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., et al. (2017). Attention is all you need. arXiv preprint arXiv:1706.03762.
Vayena E, C. I., Blasimme A. (2018).
Machine learning in medicine: Addressing ethical challenges.
PLOT Medicine,
15(11), 1–4.
https://doi.org/10.1371/journal.pmed.1002689
Wang, J., Li, M., Hu, Y., & Zhu, Y. (2009).
Comparison of hospital charge prediction models for gastric cancer patients: neural network vs. decision tree models.
BMC Health Services Research,
9(1).
https://doi.org/10.1186/1472-6963-9-161
Wang, L., Lin, Z. Q., & Wong, A. (2020a). COVID-net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest x-ray images.
Scientific Reports,
10(1), 19549.
https://doi.org/10.1038/s41598-020-76550-z
Wang, L., Lin, Z. Q., & Wong, A. (2020b). COVID-net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest x-ray images.
Scientific Reports,
10(1), 19549.
https://doi.org/10.1038/s41598-020-76550-z
Wang, L., Lin, Z. Q., & Wong, A. (2020c). COVID-net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest x-ray images.
Scientific Reports,
10(1), 19549.
https://doi.org/10.1038/s41598-020-76550-z
Wang, R., Wang, X., & Inouye, D. I. (2021).
Shapley Explanation Networks.
ICLR.
https://openreview.net/forum?id=vsU0efpivw
Wang, S., Zha, Y., Li, W., Wu, Q., Li, X., Niu, M., et al. (2020a). A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis.
European Respiratory Journal,
56(2).
https://doi.org/10.1183/13993003.00775-2020
Wang, S., Zha, Y., Li, W., Wu, Q., Li, X., Niu, M., et al. (2020b). A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis.
medRxiv.
https://doi.org/10.1101/2020.03.24.20042317
Wiens, J., Guttag, J., & Horvitz, E. (2014).
A study in transfer learning: leveraging data from multiple hospitals to enhance hospital-specific predictions.
Journal of the American Medical Informatics Association,
21(4), 699–706.
https://doi.org/10.1136/amiajnl-2013-002162
Wright, M. N., & Ziegler, A. (2016).
XGBoost: A Scalable Tree Boosting System.
SIGKDD International Conference on Knowledge Discovery and Data Mining.
https://doi.org/10.1145/2939672.2939785
Wright, M. N., & Ziegler, A. (2017c).
ranger: A fast implementation of random forests for high dimensional data in
C++ and
R.
Journal of Statistical Software,
77(1), 1–17.
https://doi.org/10.18637/jss.v077.i01
Wright, M. N., & Ziegler, A. (2017d).
ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R.
Journal of Statistical Software,
77(1), 1–17.
https://doi.org/10.18637/jss.v077.i01
Wright, M. N., & Ziegler, A. (2017a).
ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R.
Journal of Statistical Software,
77(1), 1–17.
https://doi.org/10.18637/jss.v077.i01
Wright, M. N., & Ziegler, A. (2017b).
ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R.
Journal of Statistical Software,
77(1), 1–17.
https://doi.org/10.18637/jss.v077.i01
WUoT. (2020). ML case studies: Reproducibility of scientific papers.
https://mini-pw.github.io/2020L-WB-Book/reproducibility.html
XGBoost:
A scalable tree boosting system. (2016).
CoRR,
abs/1603.02754.
http://arxiv.org/abs/1603.02754
Yan, L., Zhang, H.-T., Goncalves, J., Xiao, Y., Wang, M., Guo, Y., et al. (2020a).
An interpretable mortality prediction model for COVID-19 patients.
Nature Machine Intelligence,
2(5), 283–288.
https://doi.org/10.1038/s42256-020-0180-7
Yan, L., Zhang, H.-T., Goncalves, J., Xiao, Y., Wang, M., Guo, Y., et al. (2020b).
An interpretable mortality prediction model for COVID-19 patients.
Nature Machine Intelligence,
2(5), 283–288.
https://doi.org/10.1038/s42256-020-0180-7
Yan, L., Zhang, H.-T., Goncalves, J., Xiao, Y., Wang, M., Guo, Y., et al. (2020c).
An interpretable mortality prediction model for COVID-19 patients.
Nature Machine Intelligence,
2(5), 283--288.
https://www.nature.com/articles/s42256-020-0180-7
Yan, L., Zhang, H.-T., Goncalves, J., Xiao, Y., Wang, M., Guo, Y., et al. (2020d).
An interpretable mortality prediction model for COVID-19 patients.
Nature Machine Intelligence,
2(5), 283--288.
https://www.nature.com/articles/s42256-020-0180-7
Yildiz, B., Hung, H., Krijthe, J. H., Liem, C. C. S., Loog, M., Migut, G., et al. (2021). ReproducedPapers.org: Openly teaching and structuring machine learning reproducibility. In B. Kerautret, M. Colom, A. Krähenbühl, D. Lopresti, P. Monasse, & H. Talbot (Eds.), Reproducible research in pattern recognition (pp. 3–11). Cham: Springer International Publishing.
Yoo, Y. (2010). Computing in everyday life: A call for research on experiential computing.
MIS quarterly, 213–231.
https://doi.org/10.2307/20721425
Yu, H., Huang, X., Hu, X., & Cai, H. (2010). A comparative study on data mining algorithms for individual credit risk evaluation. In
2010 international conference on management of e-commerce and e-government (pp. 35–38). IEEE.
https://doi.org/10.1109/ICMeCG.2010.16
Zaimi Aldo, W. M., Herman, V., Antonsanti, P.-L., Perone, C. S., & Cohen-Adad, J. (2018). AxonDeepSeg: Automatic axon and myelin segmentation from microscopy data using convolutional neural networks.
Scientific Reports,
8(1), 3816.
https://doi.org/10.1038/s41598-018-22181-4
Zhao, Q., Meng, M., Kumar, R., Wu, Y., Huang, J., Deng, Y., et al. (2020).
Lymphopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A systemic review and meta-analysis.
International Journal of Infectious Diseases,
96, 131–135.
https://doi.org/10.1016/j.ijid.2020.04.086
Zhao, Y., Chetty, G., & Tran, D. (2019). Deep learning with XGBoost for real estate appraisal, 1396–1401.
https://doi.org/10.1109/SSCI44817.2019.9002790
Zheng, Y., Zhu, Y., Ji, M., Wang, R., Liu, X., Zhang, M., et al. (2020a).
A Learning-Based Model to Evaluate Hospitalization Priority in COVID-19 Pandemics.
Patterns,
1(6), 100092.
https://doi.org/10.1016/j.patter.2020.100092
Zheng, Y., Zhu, Y., Ji, M., Wang, R., Liu, X., Zhang, M., et al. (2020b).
A Learning-Based Model to Evaluate Hospitalization Priority in COVID-19 Pandemics.
Patterns,
1(9), 100173.
https://doi.org/10.1016/j.patter.2020.100173
Zhou, Z.-H., & Feng, J. (2017). Deep forest: Towards an alternative to deep neural networks.
CoRR,
abs/1702.08835.
http://arxiv.org/abs/1702.08835
Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., & Liang, J. (2019). UNet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Transactions on Medical Imaging.