1.4 Reproducibility of outdated articles about up-to-date R packages
Authors: Zuzanna Mróz, Aleksander Podsiad, Michał Wdowski (Warsaw University of Technology)
1.4.1 Abstract
The inability to reproduce scientific articles may be due to the passage of time. Factors such as changes in data or software updates - whether author-dependent or not - may make it difficult to reproduce the results after a long time. It would seem obvious that old scientific articles about R-packages are usually difficult to reproduce. But what if this package is still supported? In what way the continuous support changes the possibility of reproducing these articles? In this article we will look at examples of still updated R-packages from 2010 or older in order to analyse the type and degree of changes.
1.4.2 Introduction and Motivation
The problem of the inability to reproduce the results of research presented in a scientific article may result from a number of reasons - at each stage of design, implementation, analysis and description of research results we must remember the problem of reproducibility - without sufficient attention paid to it, there is no chance to ensure the possibility of reproducing the results obtained by one team at a later time and by other people who often do not have full knowledge of the scope presented in the article. Reproducibility is a problem in both business and science. Science, because it allows credibility of research results (McNutt 2014). Business, because we care about the correct operation of technology in any environment (Anda, Sjøberg, and Mockus 2009). As cited from “What does research reproducibility mean?” (Goodman, Fanelli, and Ioannidis 2016b); “Although the importance of multiple studies corroborating a given result is acknowledged in virtually all of the sciences, the modern use of “reproducible research” was originally applied not to corroboration, but to transparency, with application in the computational sciences. Computer scientist Jon Claerbout coined the term and associated it with a software platform and set of procedures that permit the reader of a paper to see the entire processing trail from the raw data and code to figures and tables. This concept has been carried forward into many data-intensive domains, including epidemiology, computational biology, economics, and clinical trials. According to a U.S. National Science Foundation (NSF) subcommittee on replicability in science, “reproducibility refers to the ability of a researcher to duplicate the results of a prior study using the same materials as were used by the original investigator. That is, a second researcher might use the same raw data to build the same analysis files and implement the same statistical analysis in an attempt to yield the same results…. Reproducibility is a minimum necessary condition for a finding to be believable and informative.”
1.4.4 Methodology
We have checked 13 articles with 16 R packages from at least 10 years ago to ensure that the code chunks match these categories:
- ade4: Implementing the Duality Diagram for Ecologists (Dray and Dufour 2007)
- AdMit: Adaptive Mixtures of Student-t Distributions (Ardia, Hoogerheide, and Dijk 2009)
- asympTest: A Simple R Package for Classical Parametric Statistical Tests and Confidence Intervals in Large Samples (Coeurjolly et al. 2009)
- bio.infer: Maximum Likelihood Method for Predicting Environmental Conditions from Assemblage Composition (Yuan 2007)
- deSolve, bvpSolve, ReacTran and RootSolve: R packages introducing methods of solving differential equations in R (Soetaert, Petzoldt, and Setzer 2010)
- EMD: Empirical Mode Decomposition and Hilbert Spectrum (Kim and Oh 2009)
- mvtnorm: New Numerical Algorithm for Multivariate Normal Probabilities (Mi, Miwa, and Hothorn 2009)
- neuralnet: Training of Neural Networks (Günther and Fritsch 2010)
- party: A New, Conditional Variable-Importance Measure for Random Forests Available in the party Package (Strobl, Hothorn, and Zeileis 2009)
- pls: principal Component and Partial Least Squares Regression in R (Mevik and Wehrens 2007)
- PMML: An Open Standard for Sharing Models (Guazzelli et al. 2009)
- tmvtnorm: A Package for the TruncatedMultivariate Normal Distribution (Wilhelm and Manjunath 2010)
- untb: an R Package For Simulating Ecological Drift Under the Unified Neutral Theory of Biodiversity (Hankin 2007)
All articles come from the R Journal and Journal of Statistical Software.
In our research on the subject we have decided to divide the code from the articles into chunks, according to the principle that each chunk has its own output, to which we give an evaluation according to the criteria we have set. In the process of testing and reproducing various articles, we have identified five categories, and marked them as follows:
- FULLY REP (no reproductive problems - the results were identical to original results shown in the article),
- MOSTLY REP (when the results were not ideally identical to the original, but in our opinion the chunks were working according to their purpose in the context of their article),
- HAD TO CHANGE STH (when we had to modify the code to produce correct results that will work in our current R version and can be neatly displayed in a document generated by markdown),
- HAD TO CHANGE & STILL SOMEWHAT DIFFERENT (the code had to be changed and the results were not perfect, but they were correct in the terms of the aforementioned category MOSTLY REP, but we could consider them as satisfactory),
- NO REP (no reproducibility, either due to changes through time or problems with the article that had already been there, regardless of differences in R across years).
These criteria can be considered not subjective, but setting such boundaries does not cause confusion in categorisation, thus we decided to use them in order to research and describe the introduced number of articles about R packages from at least 10 years ago.
In some of the articles we found specific types of problems:
- There was no access to data or objects referred to in later calculations,
- The results were similar to the original, but the differences were most often due to the random generation of objects. This error was usually reduced later, when the package created some kind of data summary - then the result had a very small relative error with the original result.
- The names of individual variables or some of their attributes changed (e.g. column names in the data frame).
When data or objects were unaccessible and there could not be found any alternative or history of the dataset, we classified this chunk as NO REP. What is more, in most cases chunks dependent on such objects automatically became NO REP too. However, if the code was only partially dependent on the lost dataset, it was classified as MOSTLY REP.
When we stumbled upon problems with randomly generated objects, where the values were obviously different, but after aggregating the data summaries were close to original, such chunks were marked as MOSTLY REP. As we were reproducing various articles, this has become quite significant problem that a large number of publications were struggling with.
If data could be somehow fixed - for example by changing column names, it was given HAD TO CHANGE STH mark, and the dependent chunks’ marks were not influenced by this change, which theoretically was allowing them to be marked even as FULLY REP.
1.4.5 Results
Here can be seen the in-depth report. Below are the summarised results of our research.
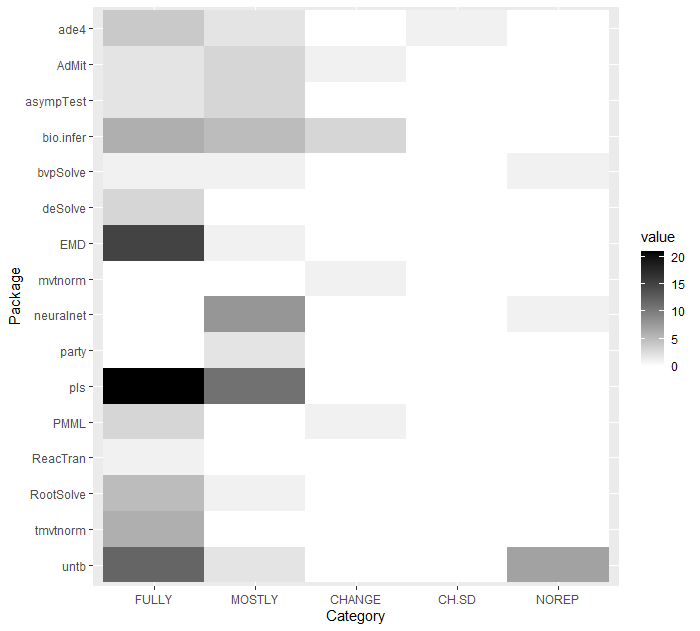
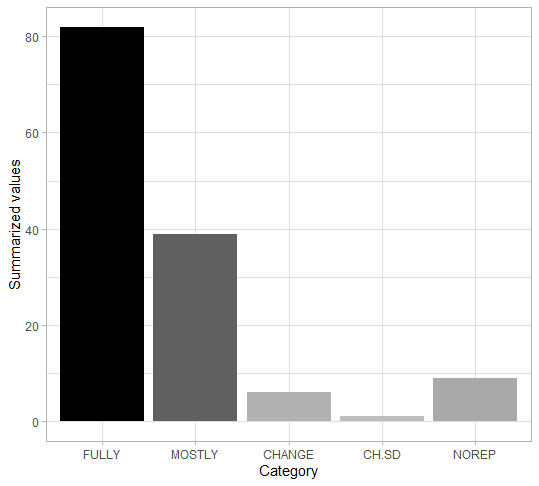
As it can be seen, the vast majority of chunks are fully reproducible. Even if the chunk is not executed identically to the original one, in most cases it differs only slightly, and the package itself serves its purpose. 88.3% (121/137) of the chunks are executed perfectly or correctly (within our subjective category of being acceptably incorrect), while 93.4% (128/137) of the chunks are working well enough not to throw errors. In practice, only 6.6% (9/137) of chunks were completely irreproducible, which would seem surprising for more-than-a-decade-old articles.
However, given that we have focused particularly on packages that are still being developed, this is quite a feasible result. This can be seen quite clearly by the percentage of the chunks that required minor changes or slightly differed from the results shown in the article - there were 33.6% (46/137), which is clearly a result of the updates or changes that occured in the ever evolving R environment. Of course during our research we stumbled upon numerous packages that have not been updated since years or that have even been deleted from CRAN repository, so they were not within our field of interest. Nonetheless, we would like to emphasize that the results should not suggest that one-decade-old articles are reproducible.
1.4.6 Summary and conclusions
To sum up, in most cases the packages we examined performed their tasks correctly. The packages themselves have of course changed, but its idea remained the same. Usually new features or improvements were added, but the idea behind the package was the same as it used to be. As a result, most of the packages still managed to cope with the problems of the old ones, in reproduction usually suffering from missing external data or unavoidable changes in the R language itself. All in all, almost in all cases the package does the job in spirit, differing from its old good ways only in vague confusion caused by neverending winds of change.
It can therefore be concluded that most packages that we’ve checked are fully backward compatible, which is good programming practice. In order to increase the reproducibility of articles, this should definitely be taken care of. Additionally, authors should include supplements to their articles, that always help you understand and copy the code.
References
Anda, Bente, Dag Sjøberg, and Audris Mockus. 2009. “Variability and Reproducibility in Software Engineering: A Study of Four Companies That Developed the Same System.” Software Engineering, IEEE Transactions on 35 (July): 407–29. https://doi.org/10.1109/TSE.2008.89.
Ardia, David, Lennart F. Hoogerheide, and Herman K. van Dijk. 2009. “AdMit.” The R Journal 1 (1): 25–30. https://doi.org/10.32614/RJ-2009-003.
Coeurjolly, J.-F., R. Drouilhet, P. Lafaye de Micheaux, and J.-F. Robineau. 2009. “asympTest: A Simple R Package for Classical Parametric Statistical Tests and Confidence Intervals in Large Samples.” The R Journal 1 (2): 26–30. https://doi.org/10.32614/RJ-2009-015.
Dray, Stéphane, and Anne-Béatrice Dufour. 2007. “The Ade4 Package: Implementing the Duality Diagram for Ecologists.” Journal of Statistical Software, Articles 22 (4): 1–20. https://doi.org/10.18637/jss.v022.i04.
Goodman, Steven N., Daniele Fanelli, and John P. A. Ioannidis. 2016a. “What Does Research Reproducibility Mean?” Science Translational Medicine 8 (341). https://doi.org/10.1126/scitranslmed.aaf5027.
2016b. “What Does Research Reproducibility Mean?” Science Translational Medicine 8 (341): 341ps12–341ps12. https://doi.org/10.1126/scitranslmed.aaf5027.Guazzelli, Alex, Michael Zeller, Wen-Ching Lin, and Graham Williams. 2009. “PMML: An Open Standard for Sharing Models.” The R Journal 1 (1): 60–65. https://doi.org/10.32614/RJ-2009-010.
Günther, Frauke, and Stefan Fritsch. 2010. “neuralnet: Training of Neural Networks.” The R Journal 2 (1): 30–38. https://doi.org/10.32614/RJ-2010-006.
Hankin, Robin. 2007. “Introducing Untb, an R Package for Simulating Ecological Drift Under the Unified Neutral Theory of Biodiversity.” Journal of Statistical Software, Articles 22 (12): 1–15. https://doi.org/10.18637/jss.v022.i12.
Kim, Donghoh, and Hee-Seok Oh. 2009. “EMD: A Package for Empirical Mode Decomposition and Hilbert Spectrum.” The R Journal 1 (1): 40–46. https://doi.org/10.32614/RJ-2009-002.
McNutt, Marcia. 2014. “Journals Unite for Reproducibility.” Science 346 (6210): 679–79. https://doi.org/10.1126/science.aaa1724.
Mevik, Björn-Helge, and Ron Wehrens. 2007. “The Pls Package: Principal Component and Partial Least Squares Regression in R.” Journal of Statistical Software, Articles 18 (2): 1–23. https://doi.org/10.18637/jss.v018.i02.
Mi, Xuefei, Tetsuhisa Miwa, and Torsten Hothorn. 2009. “New Numerical Algorithm for Multivariate Normal Probabilities in Package mvtnorm.” The R Journal 1 (1): 37–39. https://doi.org/10.32614/RJ-2009-001.
Patil, Prasad, Roger D. Peng, and Jeffrey T. Leek. 2016. “A Statistical Definition for Reproducibility and Replicability.” Science. https://doi.org/10.1101/066803.
Peng, Roger D. 2011. “Reproducible Research in Computational Science.” Science 334 (6060): 1226–7. https://doi.org/10.1126/science.1213847.
Soetaert, Karline, Thomas Petzoldt, and R. Woodrow Setzer. 2010. “Solving Differential Equations in R.” The R Journal 2 (2): 5–15. https://doi.org/10.32614/RJ-2010-013.
Strobl, Carolin, Torsten Hothorn, and Achim Zeileis. 2009. “Party on!” The R Journal 1 (2): 14–17. https://doi.org/10.32614/RJ-2009-013.
Wilhelm, Stefan, and B. G. Manjunath. 2010. “tmvtnorm: A Package for the Truncated Multivariate Normal Distribution.” The R Journal 2 (1): 25–29. https://doi.org/10.32614/RJ-2010-005.
Yuan, Lester. 2007. “Maximum Likelihood Method for Predicting Environmental Conditions from Assemblage Composition: The R Package Bio.infer.” Journal of Statistical Software, Articles 22 (3): 1–20. https://doi.org/10.18637/jss.v022.i03.