Meet Hajada!
Have you heard of the Indian mathematician Hajada? We started to think about it, having read the title of the article “The Hajada Imputation Test” - it sounded somehow familiar… But you probably haven’t had any contact with him, because not so long ago there was no such man. He was born by the authors of the test and the article, and his name comes from the first letters of their names.
So what is his test?
Hajada decided to study the effectiveness and time efficiency of various methods of dealing with missing data. He juxtaposed three simple (or even naive) methods such as deleting rows or inserting random values and three more sublime methods, including mice and missForest algorithms. The experiment consisted in using these methods on several sets of data from the famous OpenML100 collection. Hajada is not a racist about the datasets, and he chose them in order to take into account the entire demographic cross section of their datasets’ community, without discriminating against anyone – there are small and large, with lots of gaps and with almost none – a paradise for supporters of political correctness, you think. And yet, Hajada is going to COMPARE them… It’s a good thing that if he does it, he at least clearly defines the criteria. The F1 score and Matthews correlation coefficient (MCC) averaged over all selected datasets. The imputation time was an additional criterion.
What conclusions did Hajada arrive at?
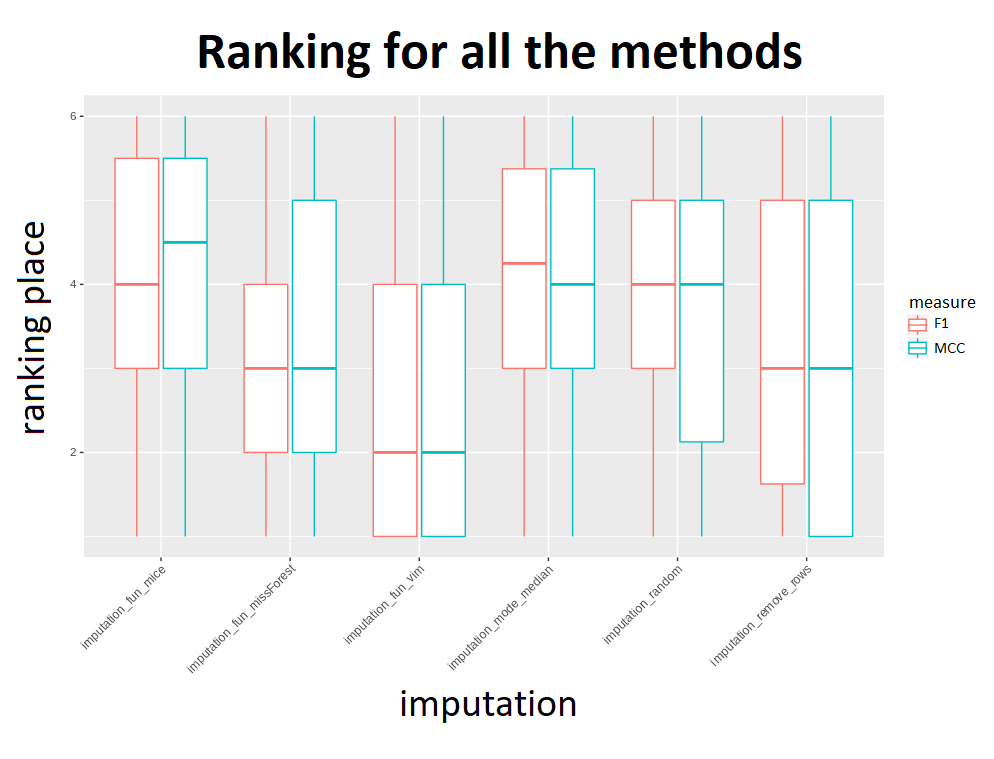
To determine which imputation method provides the best classification results Hajada created a ranking for every classification method he tested based on both measures (worth noticing – they usually resulted in the same ranks). Our concern is if a ranking for every dataset instead of a model wouldn’t also be useful in the decision process. Sadly, not every dataset was willing to cooperate and some of them caused imputation to fail, but there’s nothing surprising, hardly anyone likes to be compared… Hajada took two approaches in this case – omit the result or give it the worst rank. In both cases knn imputation from VIM package had the best average ranks. As the second best Hajada pointed out mode/median if you want fast imputation and missForest if you don’t mind waiting a little longer. Strange for us is the fact that the mode/median method had worse ranks if we take into account failed imputations because how can this method really fail? Hajada also advised against removing rows with missing values due to unstable results. In the end we know better which imputation method we will use in future but, like the article says, it is impossible to draw objective conclusions because every case is different and the results of this experiment should be confirmed on a larger number of datasets and models.
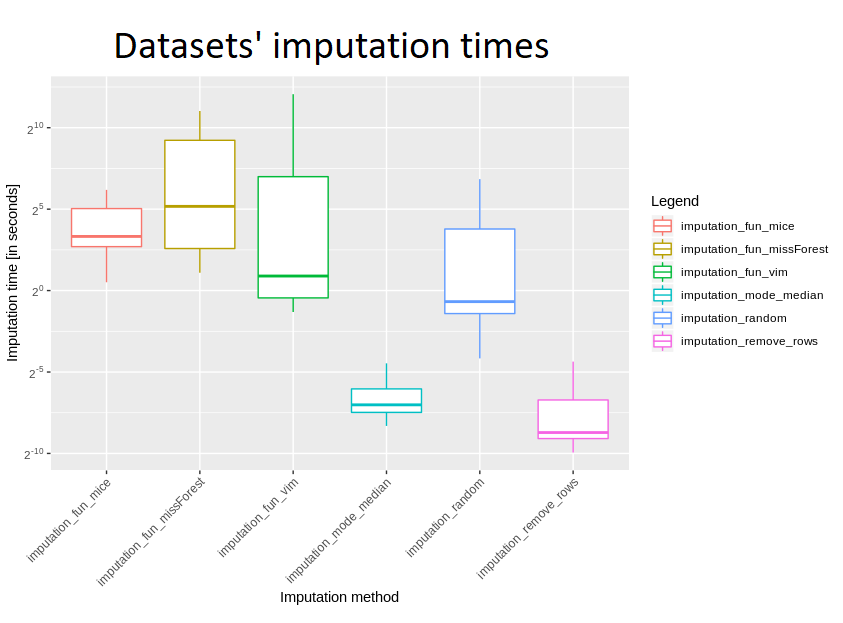
Another issue is the timing of the imputation. The most unexpected about it is that generating many random numbers is slower than computing mean/median and imputing by this method. But taking into account that we are in universe of R langugate it is very plausible. Slowness of R is almost legendary. For sure it is way slower than C and additionally it uses Mersenne twister random generator. The conclusion about time comparison is following: mice, missForest are the slowest and VIM, random number are halfway through the list. It is logical and it is worth remembering when we will have to use imputation next time. Maybe it will be worth just to use mean/median to have any imputed values and then check how other methods work. For us it was the one missing thing about time comparison. We would love to see the relationship between time of imputation and the number of rows / number of columns – it would be good to see what has a greater impact on calculation time.
What we want to say to Hajada?
Generally speaking, good work, Mr. Hajada! Now it’s time to see on your own processor how generalised these results are. There’s so much data to impute!